SMILES to IUPAC conversion with an LLM generator-scorer pattern
[mathematica llm o3 gpt4 chemistry cheminformatics Recently Andrew White tweeted about limitations of the o3-mini model to convert SMILES to IUPAC names. Here we try to correct this by using a scorer/generator LLM pattern…
For example given an input:
smiles = "CC(C)CCCC(C)C1CCC2C1(CCC3C2CC=C4C3(CCC(C4)O)C)C";
o3-mini apparently returns “lansterol”, which is incorrect. We can verify this ourselves:
LLMSynthesize[
{"I have a molecule here with the SMILES notation ", smiles,
"\n What would its IUPAC name be?"},
LLMEvaluator -> <|"Model" -> {"OpenAI", "o3-mini-2025-01-31"}|>]
(*"This structure is that of lanosterol. In full, the IUPAC name is often given as:
(3\[Beta],5\[Alpha],8\[Alpha],9\[Beta],10\[Alpha],13\[Beta],14\[Alpha],17\[Beta])-lanosta-8,24-dien-3-ol
Lanosterol is a tetracyclic triterpenoid that serves as an important biosynthetic precursor for steroids. (Note that the SMILES string shown does not include explicit stereochemical "@" markers, so the stereochemistry is assumed to be that of the naturally occurring (3\[Beta],5\[Alpha],8\[Alpha],9\[Beta],10\[Alpha],13\[Beta],14\[Alpha],17\[Beta]) isomer.)"*)
Why it is dumb to solve this problem with an LLM?
You do not really need an LLM to do this. The problem is trivially solved with the built-in MoleculeName function:
MoleculeName[smiles]
(*"epicholestrol"*)
In fact, we can query multiple sources and get different variants:
MoleculeName[smiles, #] & /@ {"PubChem", "Cactus", "Wikidata"} // TableForm
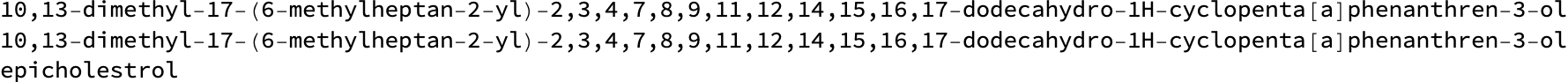
But as Prof. White notes, it is “interesting because it requires you to reason about the molecular graph (e.g., here it fails to distinguish the molecule from lanosterol). It’s like an arithmetic problem for graphs.”
Using an LLM to solve it anyway
One way to proceed might be to adopt the Multimodal Iterative LLM Solver (MILS) strategy recently introduced by Ashutosh et al. (LLMs can see and hear without any training, arXiv:2501.18096).
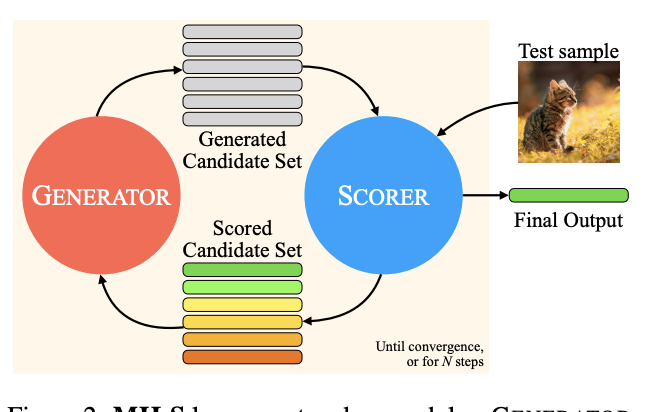 (from Figure 2 of arXiv:2501.18096)
(from Figure 2 of arXiv:2501.18096)
We define a scorer function that computes the Tanimoto similarity (of the RDKit fingerprints) between the graph specified by the query SMILES string and the graphs of the generated IUPAC names. We then iteratively use an LLM as a generator function (in this case, the Wolfram-special, which feels like gpt-4o-mini) to try to improve the results, given the past history of scored results. Prompts are just sort of made up, adapted from the Ashutosh paper appendix:
generatePrompt = StringTemplate[
"Generate 50 possible IUPAC names for the following organic molecule in SMILES notation: ``
Think step by step. Enclosed any reasoning inside of <reasoning> </reasoning> tags.
Generate 50 possible answers, enclosing each generated IUPAC name inside <answer> </answer> tags, each on its own line."];
improvePrompt = StringTemplate[
"Generate 50 possible IUPAC names for the following organic molecule in SMILES notation: ``
I am providing you with a list of previously suggested IUPAC names and scores. Higher scores means that the IUPAC name is a closer to the correct answer, but none of these are correct unless the score is 1.
``
Think step by step. Enclosed any reasoning inside of <reasoning> </reasoning> tags. Generate 50 new answers that maximize the score. Enclose each generated IUPAC name inside <answer> </answer> tags, each on its own line. Be creative and don't be afraid to come up with erroneous descriptions."];
scorer[smiles_String, candidateIUPAC_String] :=
ResourceFunction["MoleculeFingerprintSimilarity"][
Molecule[smiles], Molecule[candidateIUPAC]]
scorer[smiles_String, candidates_List] := With[
{scores = Quiet[scorer[smiles, #]& /@ candidates]},
ReverseSortBy[Last]@Pick[ Transpose[{candidates, scores}], NumericQ /@ scores]]
parse[answer_] := DeleteDuplicates@
StringCases[answer, Shortest["<answer>" ~~ x__ ~~ "</answer>"] :> x]
generator[smiles_, model_ : Automatic][{history_List, progress_List}] := With[
{result = scorer[smiles, parse[
LLMSynthesize[
improvePrompt[ smiles, Take[history, UpTo[50 ]]],
LLMEvaluator -> <|"Model" -> model|>]]]},
{DeleteDuplicatesBy[First]@ReverseSortBy[Last]@Join[history, result],
Append[
<|"round_valid" -> Length[result],
"round_best" -> result[[1, 2]],
"round_stat" -> Around[result[[All, 2]]],
"best" -> Max[result[[All, 2]], history[[All, 2]]]
|>]@progress
}]
generator[smiles_, model_ : Automatic][{}] := With[
{result = scorer[smiles, parse[
LLMSynthesize[generatePrompt@smiles,
LLMEvaluator -> <|"Model" -> model|>
]]]},
{result,
{<|"round_valid" -> Length[result],
"round_best " -> result[[1, 2]],
"round_stat" -> Around[result[[All, 2]]],
"best" -> result[[1, 2]]|>}
}]
Iterate this process 10 times with the wolfram-signature LLM:
result = Nest[ generator[smiles], {}, 10];
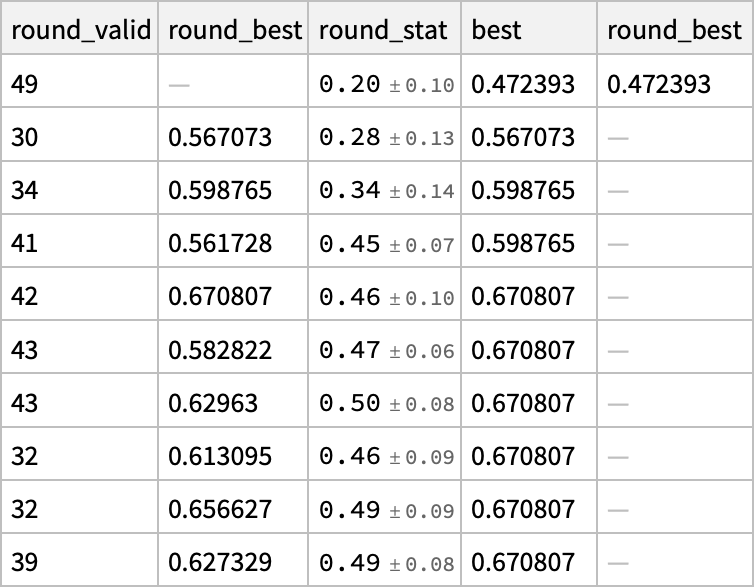
There is some improvement over iteration:
result[[2]] // Dataset
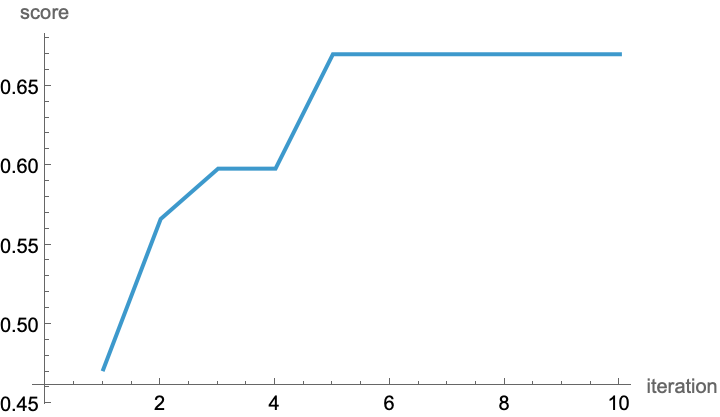
ListLinePlot[
result[[2, All, "best"]],
AxesLabel -> {"iteration", "score"}]
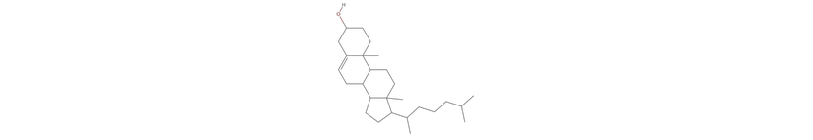
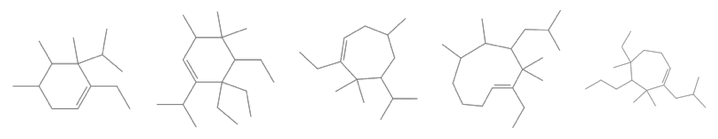
None of the top-5 candidates capture the steroid ring structure:
Thumbnail@ MoleculePlot[smiles] (* the right answer *)
Thumbnail /@ MoleculePlot /@ result[[1, 1 ;; 5, 1]] // GraphicsRow

How well does o3-mini perform?
Try it and find out (yes, we know that including “think-step-by-step” is not a best practice for reasoning models, but we are too lazy to change the prompt):
o3 = Nest[ generator[smiles, {"OpenAI", "o3-mini-2025-01-31"}], {}, 10];
o3[[2]] // Dataset
ListLinePlot[
o3[[2, All, "best"]],
AxesLabel -> {"iteration", "score"}]
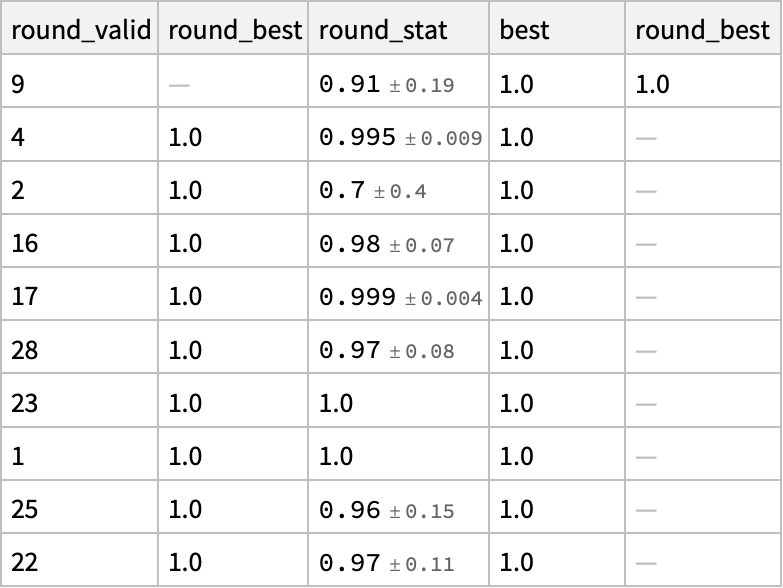
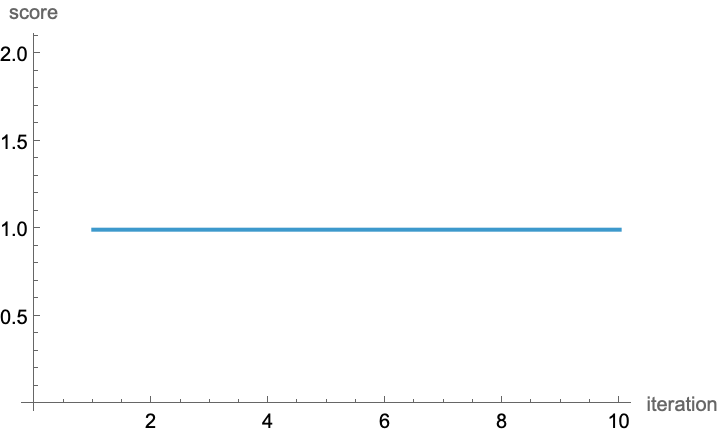
Oh! Look at that. Most of the results were wrong or even invalid, but it managed to get one right even in the first round. (I guess we didn’t have to run it for 10 iterations, should have implemented a check for convergence using NestWhile). o3 manages to get this right if given a few guesses. It is interesting to note that only 9 of the 50 proposals are actually parsed as valid molecules.
Future Ideas
-
Reinforcement fine-tuning pattern for the SMILES –> IUPAC problem
-
Can we use the generator/scorer pattern to iteratively improve explanation/rule generation by holding out a few examples and providing score information. For example, our o1 battery interface or synthesizability cases
ToJekyll["SMILES to IUPAC conversion with an LLM generator-scorer pattern",
"mathematica llm o3 gpt4 chemistry cheminformatics"]