For a Few Pixels More: A Page-Oriented Perspective on GPT-4-Vision for Scientific Data Extraction
[llm ai ml science gpt4v In a previous post, we explored some of the limitations of gpt-4-vision-preview for interpreting scientific figures in papers. A recent pre-print by Zhiling Zheng et al.1 demonstrated a useful strategy: Ask questions about images of the entire page. Surprisingly this does much better because it incorporates a variety of contextual information (figure captions, surrounding text, etc.). Here we demonstrate and adapt their strategy to solvent-solvent separation data…
As in the previous post, we import the necessary library and use a sample file. In this case we will use one of the page images we had examined before:
Import["https://raw.githubusercontent.com/antononcube/MathematicaForPrediction/master/Misc/LLMVision.m"];
SetDirectory@NotebookDirectory[];
file = "sample_pdfs/Comparison in the extraction behavior of uranium VI from nitric acid medium using CHON based extractants monoamide malonamide and diglycolamide.pdf";
exx = Import[file, "PageImages"][[6]]
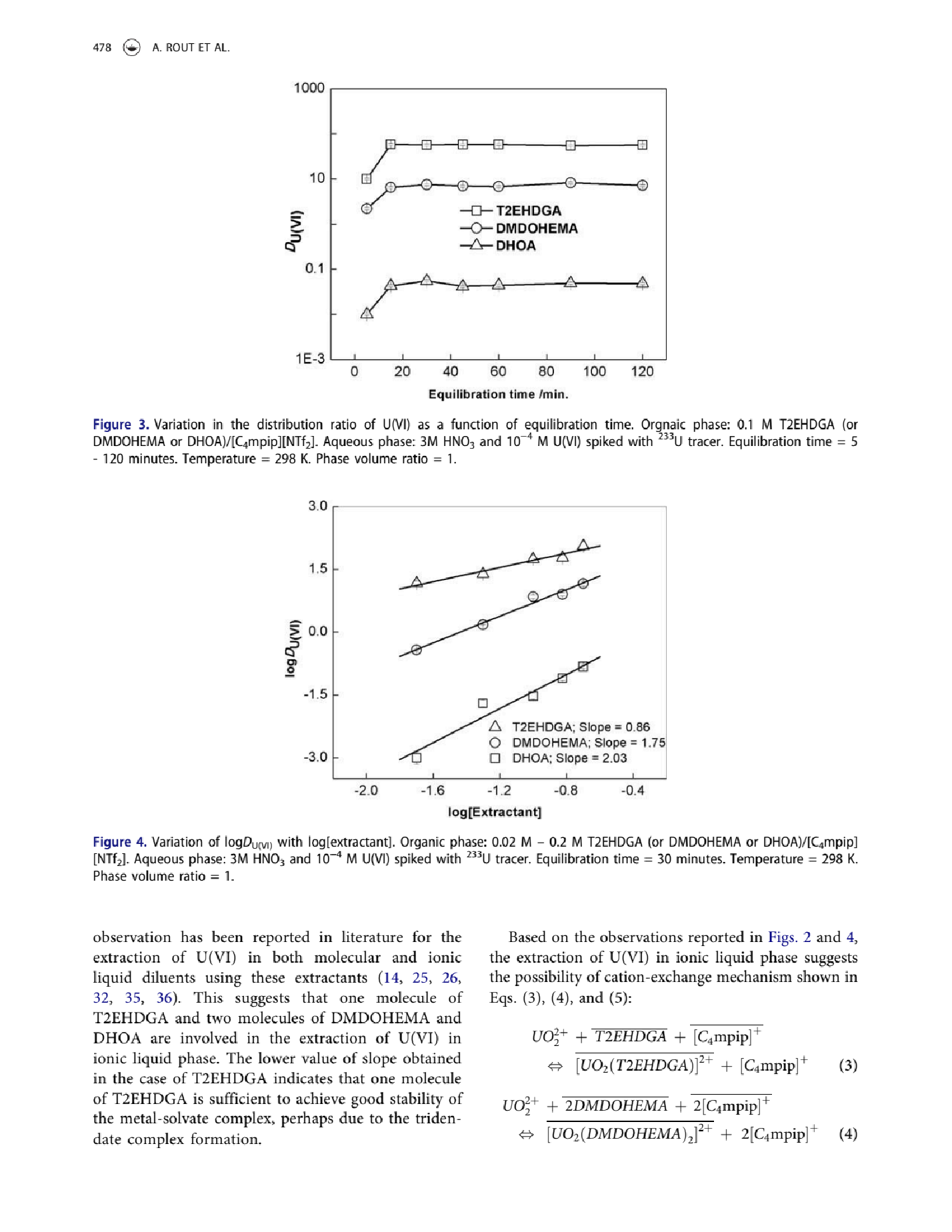
GPT-4V does a good job at identifying whether a page contains a figure that describes distribution coefficients:
LLMVisionSynthesize["Does the image contain a figure containing the distribution coefficients for a liquid-liquid separation?",
exx,
"MaxTokens" -> 600]
(*"Yes, the image includes figures (Figure 3 and Figure 4) that contain distribution coefficients for a liquid-liquid separation process. Figure 3 shows the variation in the distribution ratio of uranium(VI) as a function of equilibration time with different extractants, while Figure 4 depicts the variation of log(D_UO2) with log[Extractant]. Both figures are related to the extraction of uranium(VI) in the context of solvent extraction or liquid-liquid extraction, which is a common technique used in the separation of different components, particularly in hydrometallurgy and radioactive waste treatment."*)
The page contains information on the metal and we can extract this directly from the page image:
LLMVisionSynthesize["Does the page provide information on the metals being separated? If not provided on this page, please indicate with \"N/A\" ",
exx,
"MaxTokens" -> 600]
(*"The page you provided contains a discussion and graphical representations of the extraction of Uranium(VI) or U(VI), as indicated by the text associated with the figures and the equations presented. The figure captions mention \"spiked with ^233U tracer,\" which suggests the context is related to uranium separation or analysis. There is no explicit mention of other metals being separated in the provided text."*)
We can extract a list of the ligands being used for the separation:
LLMVisionSynthesize["Does the page provide information on the ligands being used for separations? If not provided on this page, please indicate with \"N/A\" ",
exx,
"MaxTokens" -> 600]
(*"The page contains information on the ligands being used for separations. Specifically, the ligands mentioned are T2EHDGA (N,N,N',N'-Tetra(2-ethylhexyl)diglycolamic acid), DMDODHEMA (N,N'-Dimethyl-N,N'-dioctylhexylethoxymalonamide), and DHOA (Di(2-ethylhexyl)orthophosphoric acid). In Figure 3, the distribution ratio of uranium(VI) (U(VI)) is presented as a function of equilibration time using these ligands, and in Figure 4, the variation of log D_UO2 with log[extractant] using these ligands is shown."*)
Solvent information is not quite present on this page:
LLMVisionSynthesize["Does the page provide information on the solvents being used for separations? If not provided on this page, please indicate with \"N/A\" ",
exx,
"MaxTokens" -> 600]
(*"The page provided does contain information about the solvents used for separations. In Figure 3, it specifies that the organic phase is 0.1 M T2EHDGA (or DMDOHEMA or DHOA)/[Cmim][NTf2]. For the aqueous phase, it mentions 3 M HNO3 and 10^-4 M U(VI) spiked with ^233U tracer. The caption of Figure 4 also mentions the organic phase containing 0.02 M \[Dash] 0.2 M T2EHDGA (or DMDOHEMA or DHOA)/[Cmim][NTf2]."*)
We can extract some information about reaction conditions?
LLMVisionSynthesize["Does the page provide information on the pH or acid concentration being used for separations? If not provided on this page, please indicate with \"N/A\" ",
exx,
"MaxTokens" -> 600]
(*"The page provides information on the pH indirectly by mentioning the composition of the aqueous phase. It states that the aqueous phase contains 3 M HNO3, which is a strong acid. The presence of 3 M HNO3 suggests that the aqueous solution is highly acidic; however, the exact pH value is not provided on this page. Typically, a 3 M HNO3 solution would have a pH well below 1, but if you need the precise pH, you would need to refer to additional information not provided on this page. Therefore, \"N/A\" for the exact pH value."*)
Quantitative extraction is still poor (as discussed above):
LLMVisionSynthesize["In Figure 3, identify the range of measured distribution coefficients for T2EHDGA. Report the values in a range of the y values given in the plot next to the y-axis which can be read (e.g., 0.5 - 3). If you are unsure about your answer, please indicate with \"I do not know.\"",
exx,
"MaxTokens" -> 600]
(*"The graph in Figure 3 shows the variation in the distribution ratio of U(VI) for different extractants over equilibration time. Looking at the plot for T2EHDGA, which is represented by squares, the distribution coefficients (D_UO2^VI) appear to remain relatively stable over the time measured. The y-axis represents the distribution ratio on a logarithmic scale.For T2EHDGA, the values of the distribution ratio range between approximately 1 and 10 throughout the duration of the experiment, as indicated by the positioning of the squares between these two values on the y-axis. If you need a more precise range within this bracket, I am unable to provide a more accurate number due to the resolution of the graph. However, given what is visible, you can report that the range of measured distribution coefficients for T2EHDGA lies approximately between 1 and 10."*)
Telling the model that the scale is a log-scale does not seem to improve the results:
LLMVisionSynthesize["In Figure 3, identify the range of measured distribution coefficients for T2EHDGA. Report the values in a range of the y values given in the plot next to the y-axis which can be read (e.g., 1 - 3). Note that the y-axis is on a logarithmic scale. If you are unsure about your answer, please indicate with \"I do not know.\"",
exx,
"MaxTokens" -> 600]
(*"The y-axis in Figure 3 represents the distribution ratio of U(VI), denoted as D_U(VI), and is on a logarithmic scale. For the extractant T2EHDGA, the distribution coefficient values appear to be quite stable across the different equilibration times. The range of the measured distribution coefficients for T2EHDGA spans from approximately 2 to 10, as can be observed on the y-axis next to the corresponding data points. However, without more information or a clearer image, providing an exact range can be difficult. The estimate is made based on the plotted data points in Figure 3 for T2EHDGA and the corresponding values on the y-axis."*)
Identifying ranges appears to do better in linear scale plots. Again, it is still approximative, and in the latter cases it appears to do quite poorly:
LLMVisionSynthesize["In Figure 4, identify the range of measured distribution coefficients for T2EHDGA. Report the values in a range of the y values given in the plot next to the y-axis which can be read (e.g., 1 - 3). If you are unsure about your answer, please indicate with \"I do not know.\"",
exx,
"MaxTokens" -> 600]
(*"In Figure 4 of the provided image, the y-axis represents the log of the distribution coefficient (logD_UO2(VI)), and the range of measured distribution coefficients for T2EHDGA can be identified by the region covered by the data points marked with triangles (\[CapitalDelta]).The lowest point on the T2EHDGA data series appears to be at approximately 0.5 on the y-axis, and the highest point appears to be just below 2.5. Therefore, we can report the range of logD_UO2(VI) for T2EHDGA as roughly 0.5 to just below 2.5. However, without a higher resolution image or precise data, it is difficult to provide an exact maximum value for the y-axis data for T2EHDGA. Thus, the values are approximate and based on the visual estimation from the provided plot."*)
LLMVisionSynthesize["In Figure 4, identify the range of measured distribution coefficients for DHOA. Report the values in a range of the y values given in the plot next to the y-axis which can be read (e.g., 1 - 3). If you are unsure about your answer, please indicate with \"I do not know.\"",
exx,
"MaxTokens" -> 600]
(*"In Figure 4, the measured distribution coefficients for DHOA (marked with triangles in the graph) appear to range approximately between 0.1 and 1.0 on the y-axis (logD_UO2). This range can be estimated from the points plotted for DHOA, which are positioned along the y-axis values that correspond with the logarithm scale provided."*)
However, we can successfully extract information about the types of variations in the presented data:
LLMVisionSynthesize["Does any information on this page indicate that the experiments were performed measuring the distribution coefficient at different temperatures? If you are unsure about your answer, please indicate with \"I do not know\"",
exx,
"MaxTokens" -> 500]
(*"Based on the information provided on this page, the experiments outlined in Figure 3 and Figure 4 mention a specific temperature of 298 K, which corresponds to approximately 25DegreeC. There is no indication on this page that the experiments were performed at different temperatures. The temperature is mentioned as a constant condition for each of the experiments represented in these figures. Therefore, there is no evidence here of measurements being taken at various temperatures."*)
LLMVisionSynthesize["Does any information on this page indicate that the experiments were performed measuring the distribution coefficient at different times? If you are unsure about your answer, please indicate with \"I do not know\"",
exx,
"MaxTokens" -> 500]
(*"Yes, the information provided in Figure 3 of the provided page does indicate that experiments were performed measuring the distribution coefficient (D_U(VI)) at different times. According to the caption of Figure 3, the distribution ratio of U(VI) is plotted as a function of equilibration time. The equilibration times mentioned in the caption are from 5 to 120 minutes, and the x-axis of the graph in Figure 3 displays time in minutes, which corresponds with the equilibration time described. This implies that the distribution coefficient was indeed measured at various times within the stated range. Temperature is noted to be 298 K and the phase volume ratio is given as 1."*)
How well can we extract locations? tl;dr–not well. We get pretty inconsistent results. Probably best to leave this out.
LLMVisionSynthesize["Localize the region in this page containing a distribution coefficient plot and its figure caption. Regions are represented by [ x1, y1, x2, y2 ] coordinates. x1 and x2 are the leftmost and rightmost positions, normalized into 0 to 1, where 0 is the left and 1 is the right. y1 and y2 are the topmost and bottommost positions, normalized into 0 to 1, where 0 is the top and 1 is the bottom. The selection should contain the entire figure and its caption. If two figures of distribution coefficients are present, return two lists specifying each figure, e.g., [ [region 1], [region 2] ]. Return only the list values.",
exx,
"MaxTokens" -> 500]
(*"[[0.08, 0.51, 0.92, 0.79]]"*)
{w, d} = ImageDimensions[exx];
{x1, y1, x2, y2} = ImportString["[0.08, 0.51, 0.92, 0.79]", "JSON"];
Thumbnail[#, Medium] &@ImageTake[exx, {y1*d, y2*d}, {x1*w, x2*w}]
Putting it together:
LLMVisionSynthesize["Please analyze the provided image, which is a page from a scientific paper, and answer the following questions:1. Does the image contain one or more figures containing the distribution coefficients for a liquid-liquid separation? Return \"true\" or \"false\" only.2. If the figure contains a distribution coefficient, in which figure (e.g., Figure 1, 2, Figure S4) is it displayed? Note that we are only interested in distribution coefficient measurements. Return the values as a JSON list (e.g., [1, 2, \"S4\"]). If not present on this page, please indicate with an empty list \"[]\".3. If distribution coefficient measurements are present, can you identify the name of the extractant ligand compound being measured? Return the values as a JSON list, (e.g., [\"DIHEBA\", \"TODGA\"]), containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\".4. If distribution coefficients are measured, what metal elements are being measured? Return the values as a JSON list (e.g., [\"Ca\", \"Bk\"]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\".5. Are there variations with respect to temperature that are measured? Return the values as a JSON list (e.g., [\"22 C\"]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\"6. Are there variations with respect to time that are measured? Return the values as a JSON list (e.g., [\"10 minutes\"]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\"7. What solvents are used in the experiments? Return the values as a JSON list (e.g., [\"toluene\"]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\"8. What pH is specified in the experiments? Return the values as a JSON list (e.g., [1.0, 5.0]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\"9. What acid concentrations are specified in the experiments? Return the values as a JSON list (e.g., [\"1 M\"]) containing any values that are present. If not provided on this page, please indicate with an empty list \"[]\"Please strictly adhere to the templates provided and return the result as a JSON dictionary. ****Template for your answer if no distribution coefficients are reported:{\"pageHasD\":false}Template for your answer if distribution coefficient information is reported:{\"page_has_D\":true, \"figures\": [Answer: from question 2], \"extractants\": [Answer: from question 3],\"metals\": [Answer: from question 4],\"temperatures\": [Answer: from question 5],\"times\": [Answer: from question 6],\"solvents\": [Answer: from question 7],\"pH\": [Answer: from question 8],\"acid_concentration\": [Answer: from question 9] }",
exx, "MaxTokens" -> 500]
(*"```json{\"page_has_D\":true, \"figures\": [3, 4], \"extractants\": [\"T2EHDGA\", \"DMDOHEMA\", \"DHOA\"],\"metals\": [\"U\"],\"temperatures\": [\"298 K\"],\"times\": [\"5 - 120 minutes\", \"30 minutes\"],\"solvents\": [\"HNO3\"],\"pH\": [],\"acid_concentration\": [\"3 M\"] }```"*)
What should we name this post?
GPT4> What are the names of Clint Eastwood cowboy movies? Ideally I want to make a pun related to GPT, AI and Vision
Clint Eastwood is known for his roles in several popular western films. Let’s gather a list of these movies.
 From the web search, we have some names of Clint Eastwood’s western movies:
From the web search, we have some names of Clint Eastwood’s western movies:
- The Good, The Bad And The Ugly
- Unforgiven
- For A Few Dollars More
- The Outlaw Josey Wales
- Paint Your Wagon
We can use these titles to create a pun related to GPT, AI, and Vision. Here are a few suggestions:
- “The Good, the Bot, and the Ugly: A GPT’s journey to AI Vision”
- “AI Vision: For a Few Pixels More”
- “Unforgiven Errors: When GPT lost its AI Vision”
- “The Outlaw Code-y Wales: A tale of GPT and AI Vision in the Wild West of Data”
- “Paint Your Wagon with Data: GPT and AI Vision’s Masterpiece”
Remember, the key to a good pun is playing with the words in a humorous or unexpected way. I hope these suggestions bring a smile to your face!
ToJekyll["For a Few Pixels More: A Page-Oriented Perspective on GPT-4-Vision for Scientific Data Extraction",
"llm ai ml science"]
Parerga and paralipomena
- Set of Mark prompting—the idea is that you add bounding boxes/infills/marks to your image before providing it as input to GPT-4V. Apparently this greatly improves performance. You can also just number all the items (and optionally provide masking or bounding boxes) and it greatly improves results and reduces hallucinations.
- Came across this in Ju Li’s CREST preprint, where they use this to adjust settings for an SEM image.
- But as a redditor said, “combine it with Meta’s segment anything and profit”
- For quick hackery, just use the YOLO v8 on the network repository
- There is some work on figure-specific extraction:
- LineEx: Data Extraction from Scientific Line Charts
- MatGB: Materials Graph Digitizer-YOLOv8x model fine tuned to segment images and label line-chart/not and then extract data from the line charts