Parsing Molecular Identifiers From the Ideal Database, part 3
[mathematica chemdraw science gpt3.5 In our last episode, we were left with 25 cases where the inferred molecule did not agree with the reported formula or molar mass, in our quest to to turn the IDEaL database into a comprehensive f-element separation database. Here we fix them by hand and generate the final result…
Import the data
SetDirectory@NotebookDirectory[];
missing = Import["2023.10.16_missing_entries_needs_proofing.xlsx", {"Dataset", 1}, "HeaderLines" -> 1]
correct = Dataset[{}];
excluded = Dataset[{}];
(*revised from last post*)
outputData[oldRecord_, mol_Molecule] := With[
{reportedMass = Interpreter[Number]@ oldRecord["reported_mass"],
computedMass = QuantityMagnitude[mol["MolarMass"]]},
Dataset@Association[
"url" -> oldRecord["url"],
"abbreviation" -> oldRecord["abbreviation"],
"SMILES" -> mol["CanonicalSMILES"],
"InChI" -> mol["InChI"]["ExternalID"],
"InChIKey" -> mol["InChIKey"]["ExternalID"],
"computed_formula" -> mol["MolecularFormulaString"],
"reported_formula" -> oldRecord["reported_formula"],
"computed_mass" -> computedMass,
"reported_mass" -> reportedMass,
"formula_matchQ" -> StringMatchQ[mol["MolecularFormulaString"], oldRecord["reported_formula"]],
"mass_matchQ" -> (Round[reportedMass] == Round[computedMass])
]]
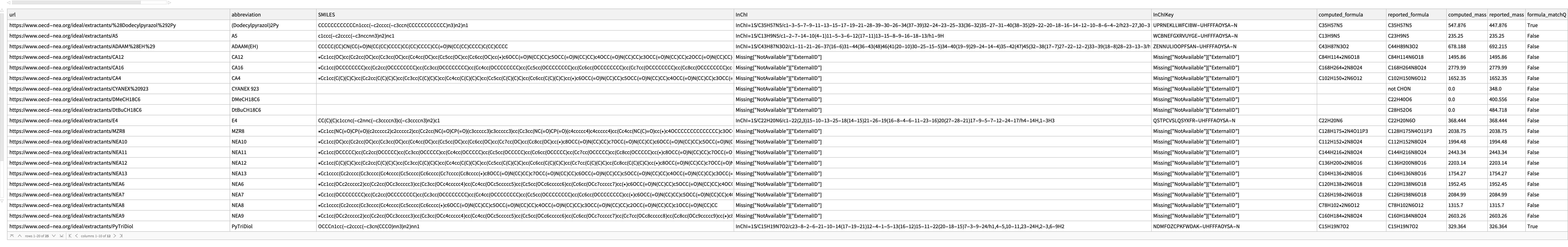
1
missing[[1]]
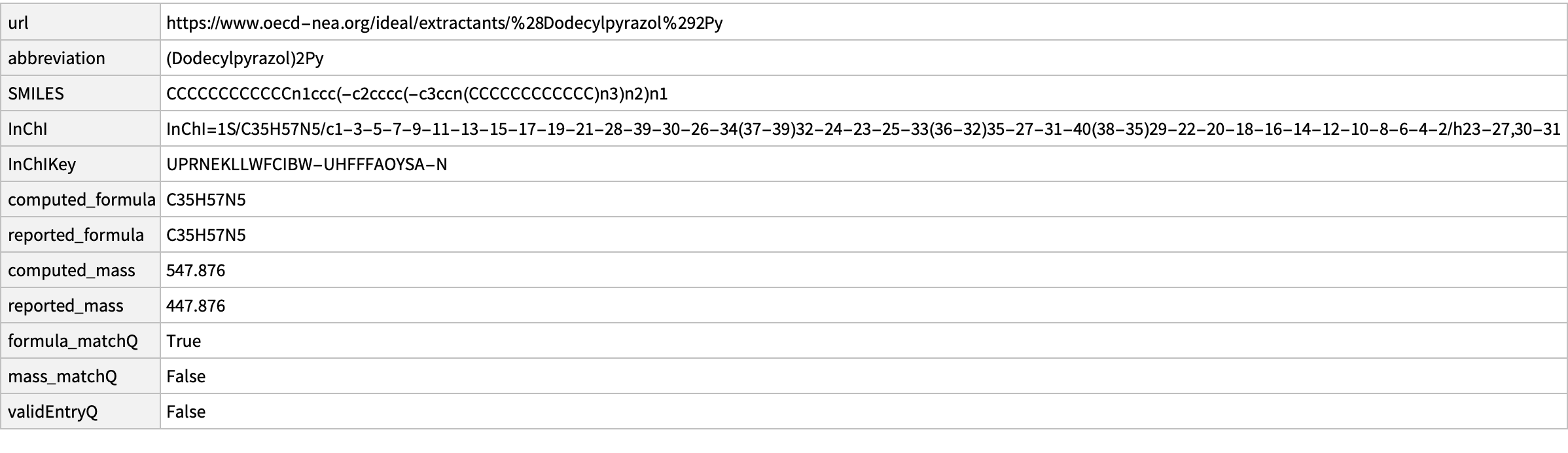
WebImage@missing[[1, "url"]]
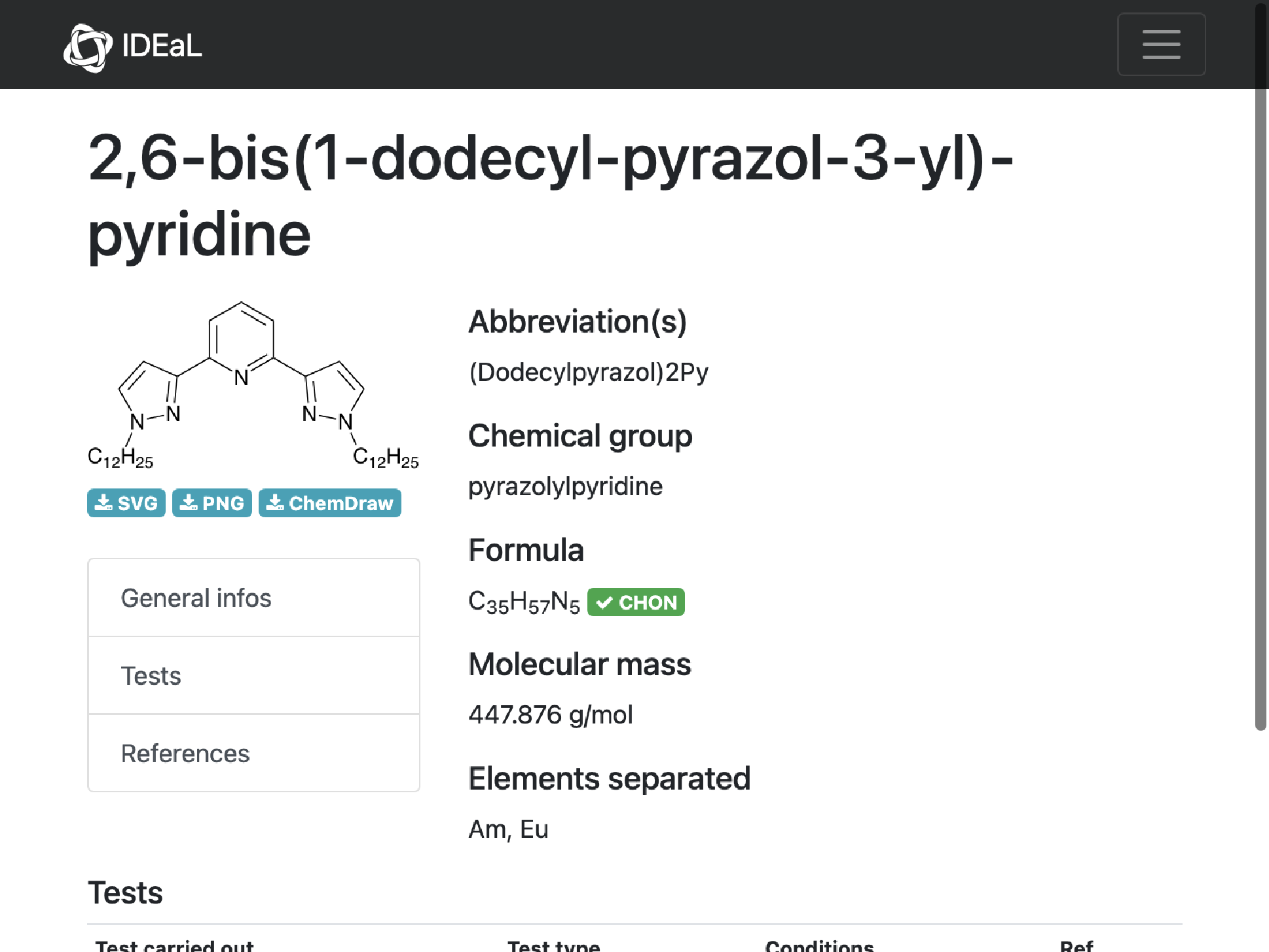
MoleculePlot@Molecule@missing[[1, "InChI"]]
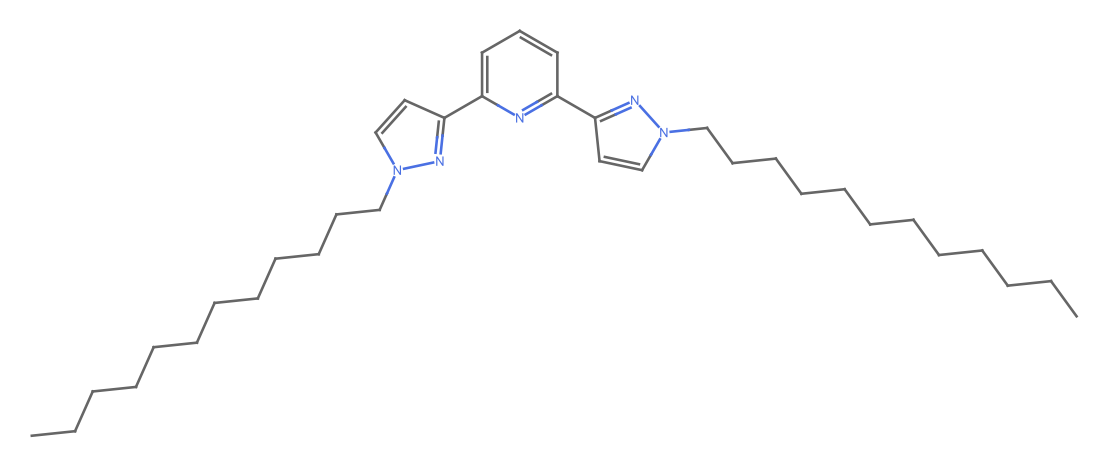
Comment: Imported structure is consistent with diagram and consistent with molecular formula. This implies an error of IDEaL mass.
AppendTo[correct, missing[[1]]]

2
missing[[2]]
WebImage@%["url"]
MoleculePlot@Molecule@%%["InChI"]
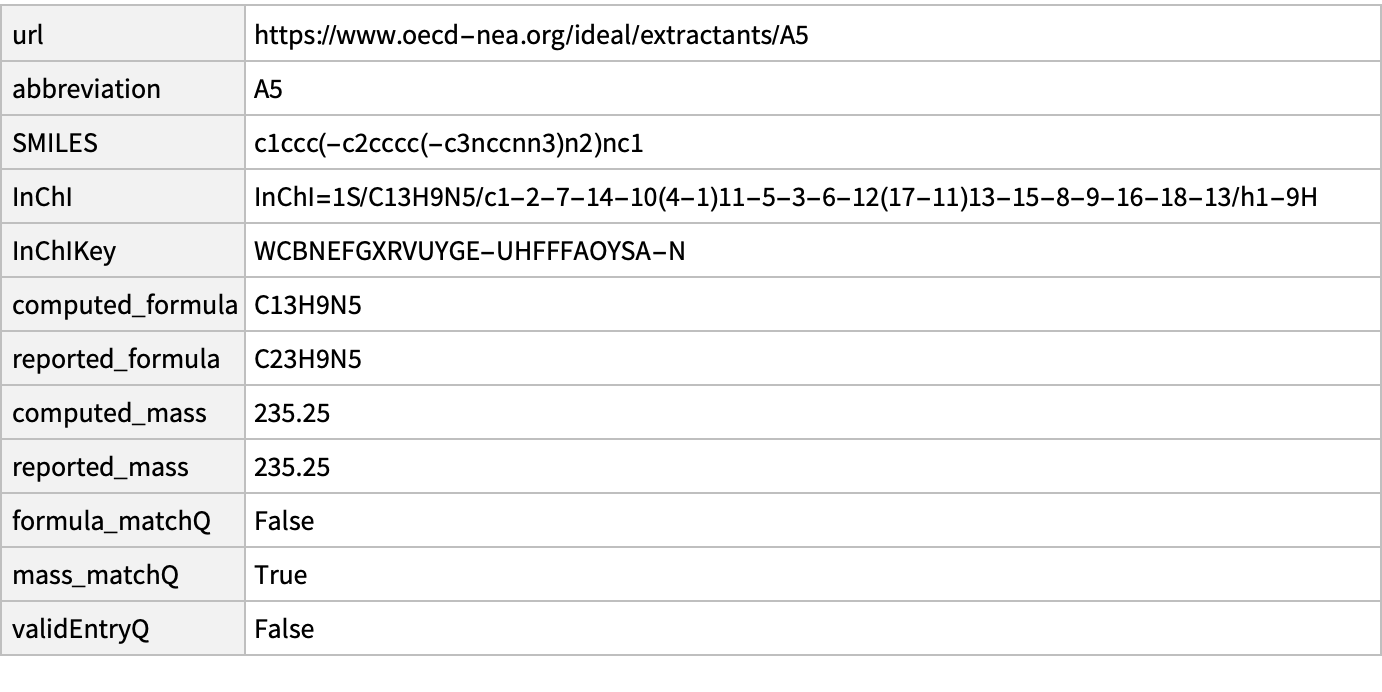
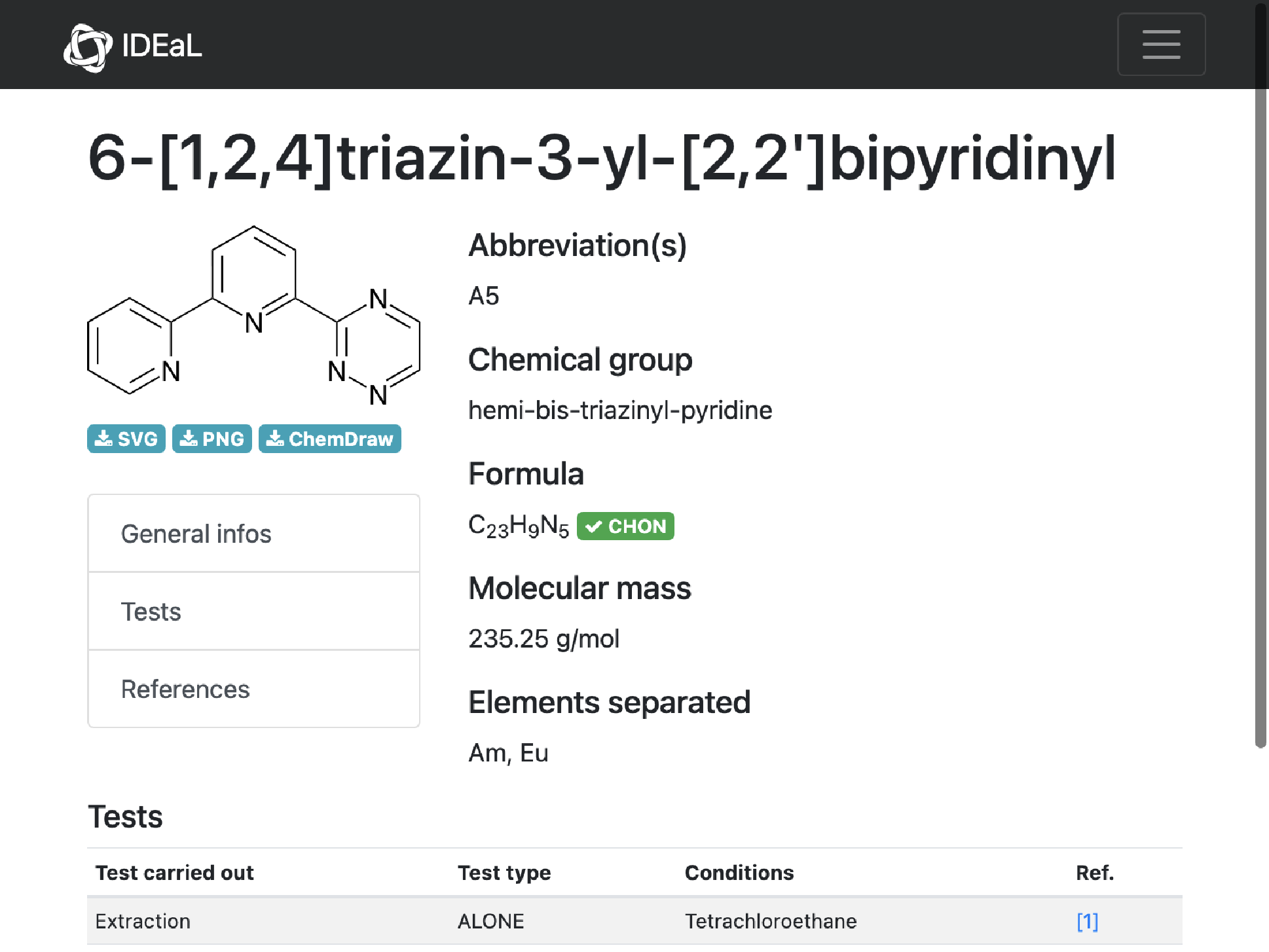
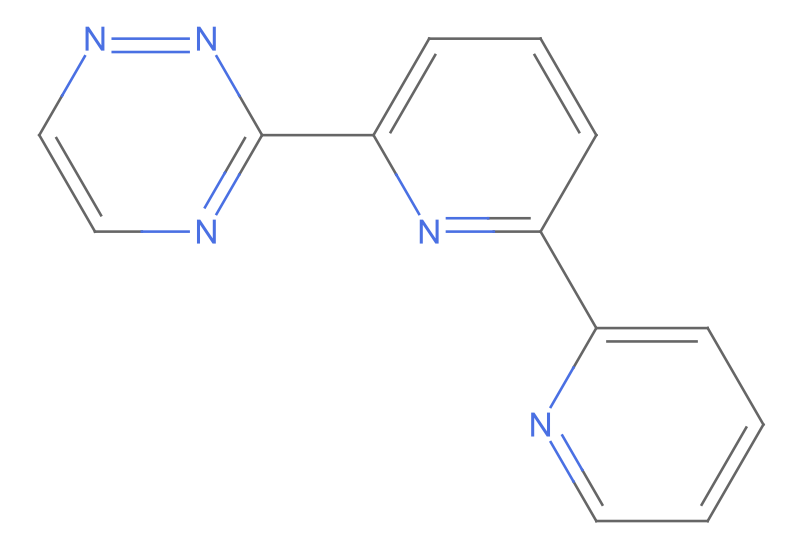
Mass and structures match. Implies error on IDEaL chemical formula
AppendTo[correct, missing[[2]]]

3
missing[[3]]
WebImage@%["url"]
MoleculePlot@Molecule@%%["InChI"]

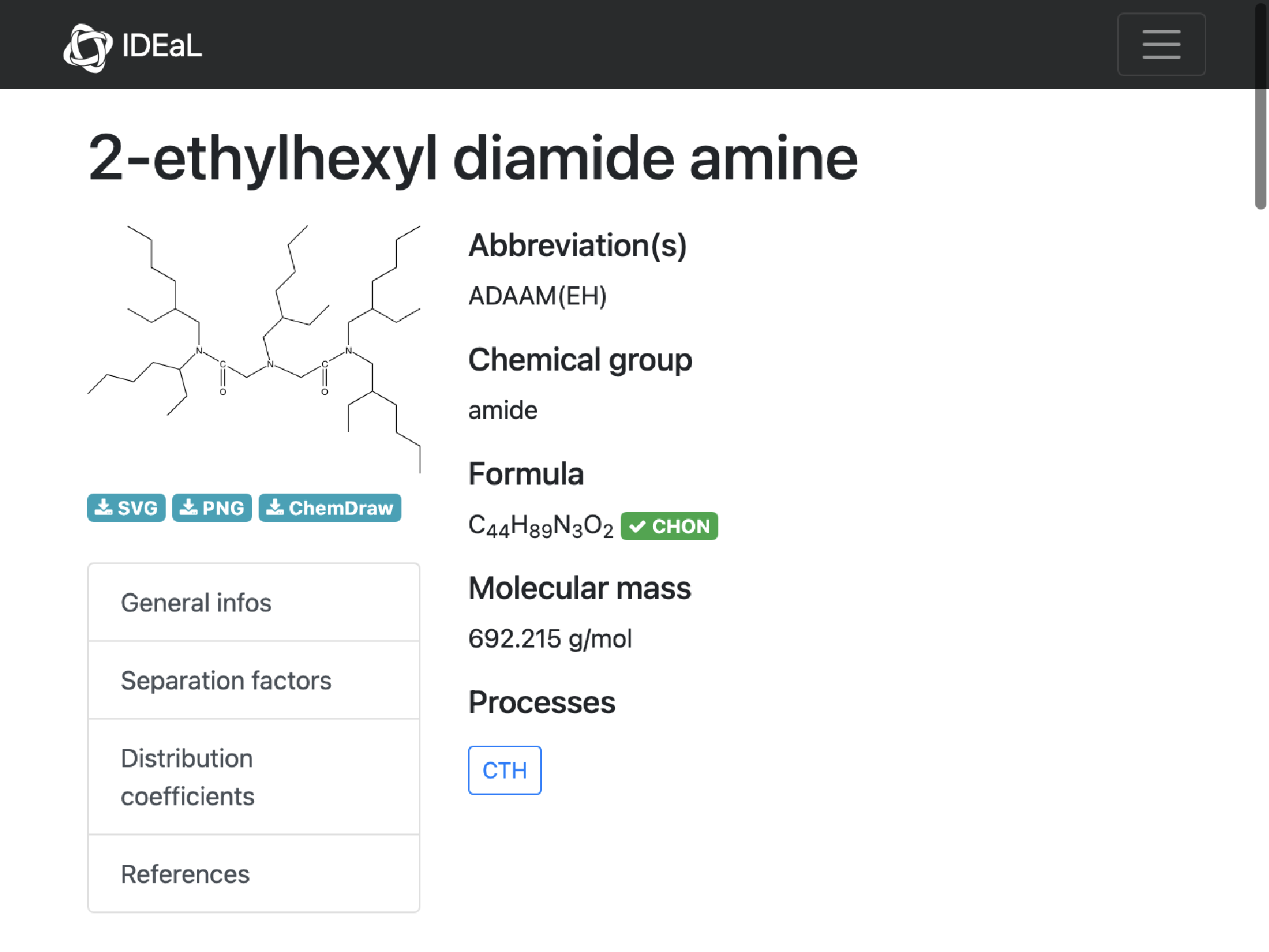
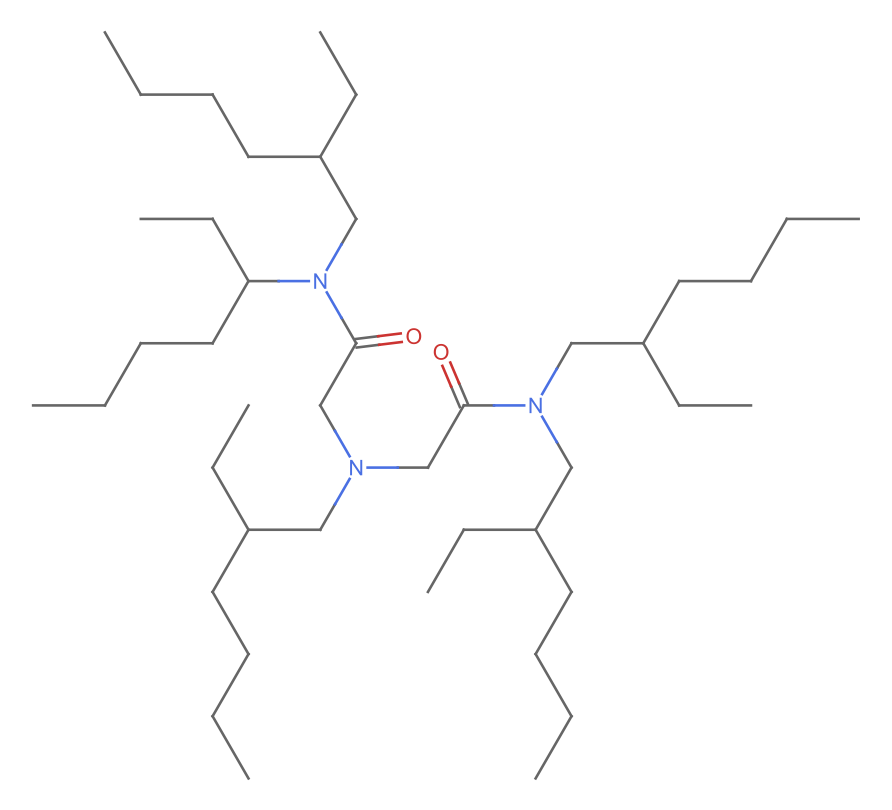
Comment: It seems strange (and unlikely) to me that one of the sidechains is different (missing a carbon compared to the others; all the others are 2-ethylhexl as implied by the name). I suspect that the formula is correct, but the structure drawn is incorrect.
CopyToClipboard@missing[[3]]["InChI"]
AppendTo[
correct,
outputData[missing[[3]], Molecule["CCCCC(CN(CC(N(CC(CCCC)CC)CC(CCCC)CC)=O)CC(N(CC(CCCC)CC)CC(CCCC)CC)=O)CC"]]]

4
missing[[4]]
WebImage@%["url"]
MoleculePlot@Molecule@%%["InChI"]

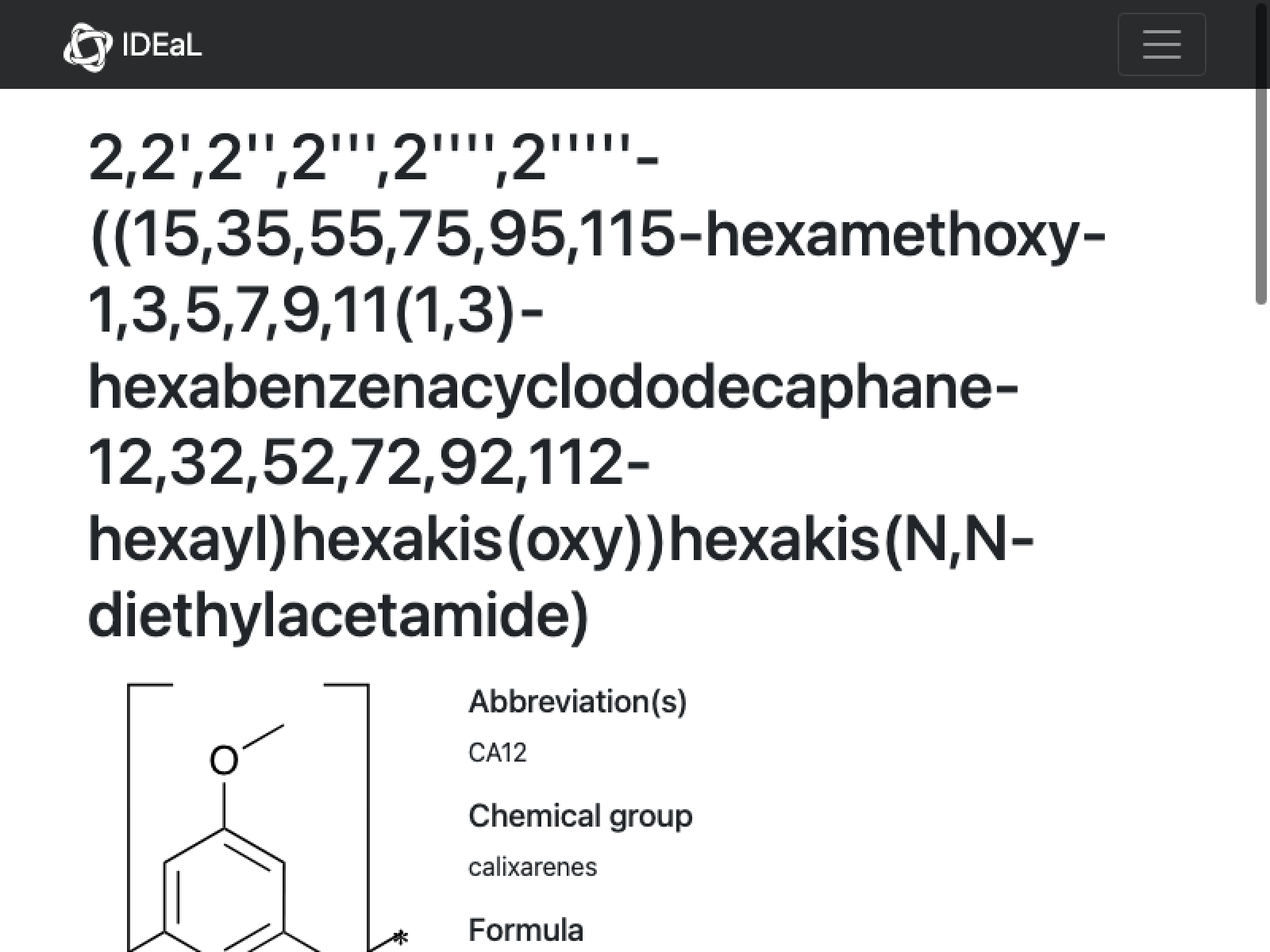


(*MoleculePlot[Molecule["Missing[\"NotAvailable\"][\"ExternalID\"]"]]*)
Comment: Looks like the structure is fine (matches molecular weight, etc.) and the only problem is that there are some extra asterisks. This is actually a six membered ring(!) We have to open this in ChemDraw and manually edit it (copying back the result)
CopyToClipboard@missing[[4]]["SMILES"]
Editing fixes the agreement…
outputData[missing[[4]], Molecule["COc1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(C6)cc(OC)c5)cc(OC)c4)cc(OC)c3)cc(OC)c2)c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c6cc(OC)c7)c1"]]
AppendTo[correct, %]


Comment: It appears we have fixed it!
5
missing[[5]]

Looks like another ring… copy to clipboard and repair
CopyToClipboard@missing[[5]]["SMILES"]
outputData[missing[[5]], Molecule["CCCCCCCCOc1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(Cc8c(OCC(N(CC)CC)=O)c(C9)cc(OCCCCCCCC)c8)cc(OCCCCCCCC)c7)cc(OCCCCCCCC)c6)cc(OCCCCCCCC)c5)cc(OCCCCCCCC)c4)cc(OCCCCCCCC)c3)cc(OCCCCCCCC)c2)c(OCC(N(CC)CC)=O)c9c1"]]
AppendTo[correct, %]


6
missing[[6]]

Comment: This appears to be a batter with all of these CA* extractants… OK
CopyToClipboard@missing[[6, "SMILES"]]
outputData[missing[[6]], Molecule["CC(C)(C)c1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(C7)cc(C(C)(C)C)c6)cc(C(C)(C)C)c5)cc(C(C)(C)C)c4)cc(C(C)(C)C)c3)cc(C(C)(C)C)c2)c(OCC(N(CC)CC)=O)c7c1"]]
AppendTo[correct, %];

7
missing[[7]]

WebImage[missing[[7, "url"]]]
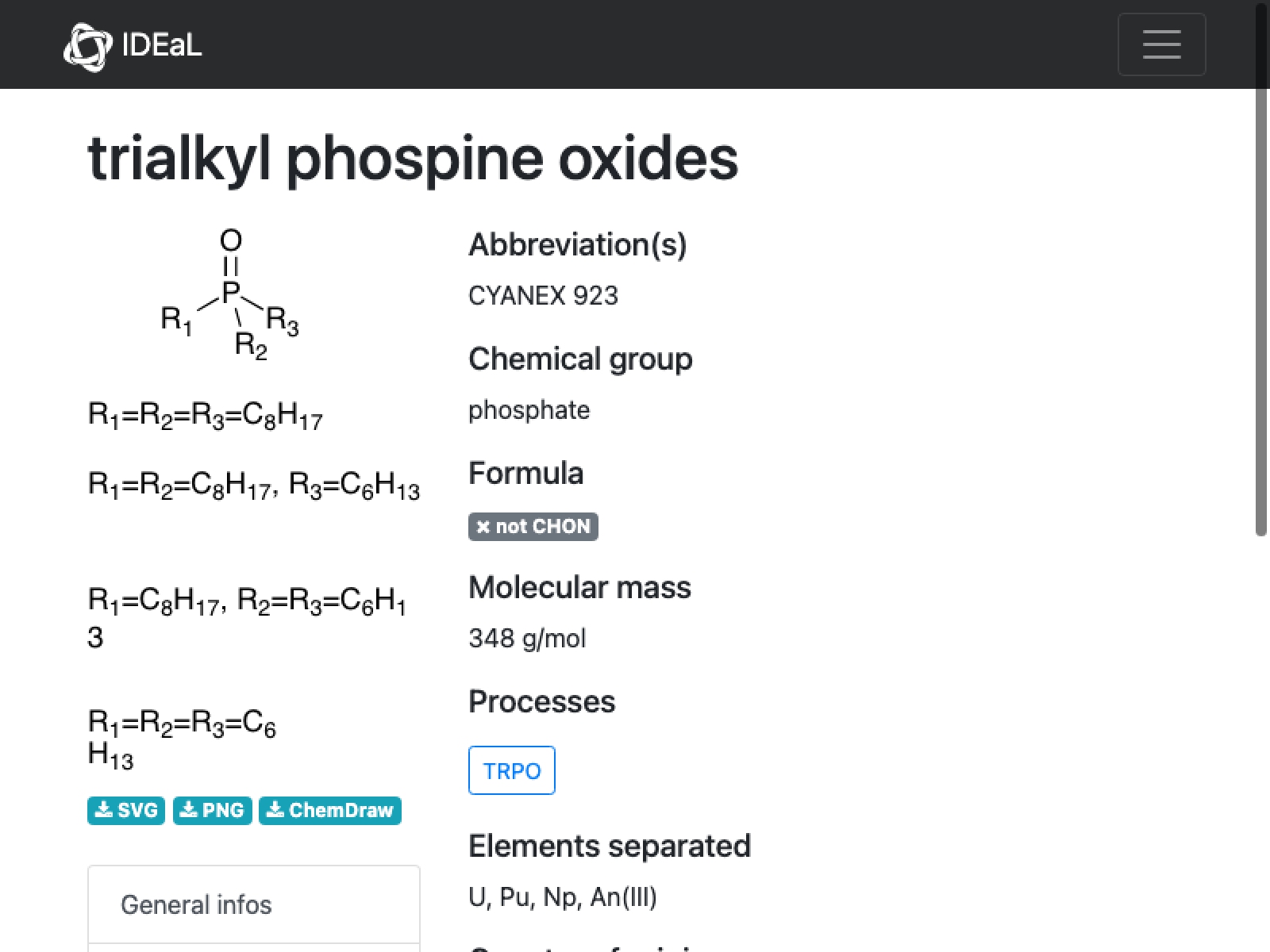
Comment: This is not a well-defined compound (it appears to be some mix?) and there are no D-values, so we are going to leave this one out….
AppendTo[excluded, missing[[7]]]

8
missing[[8]]

WebImage@missing[[8, "url"]]
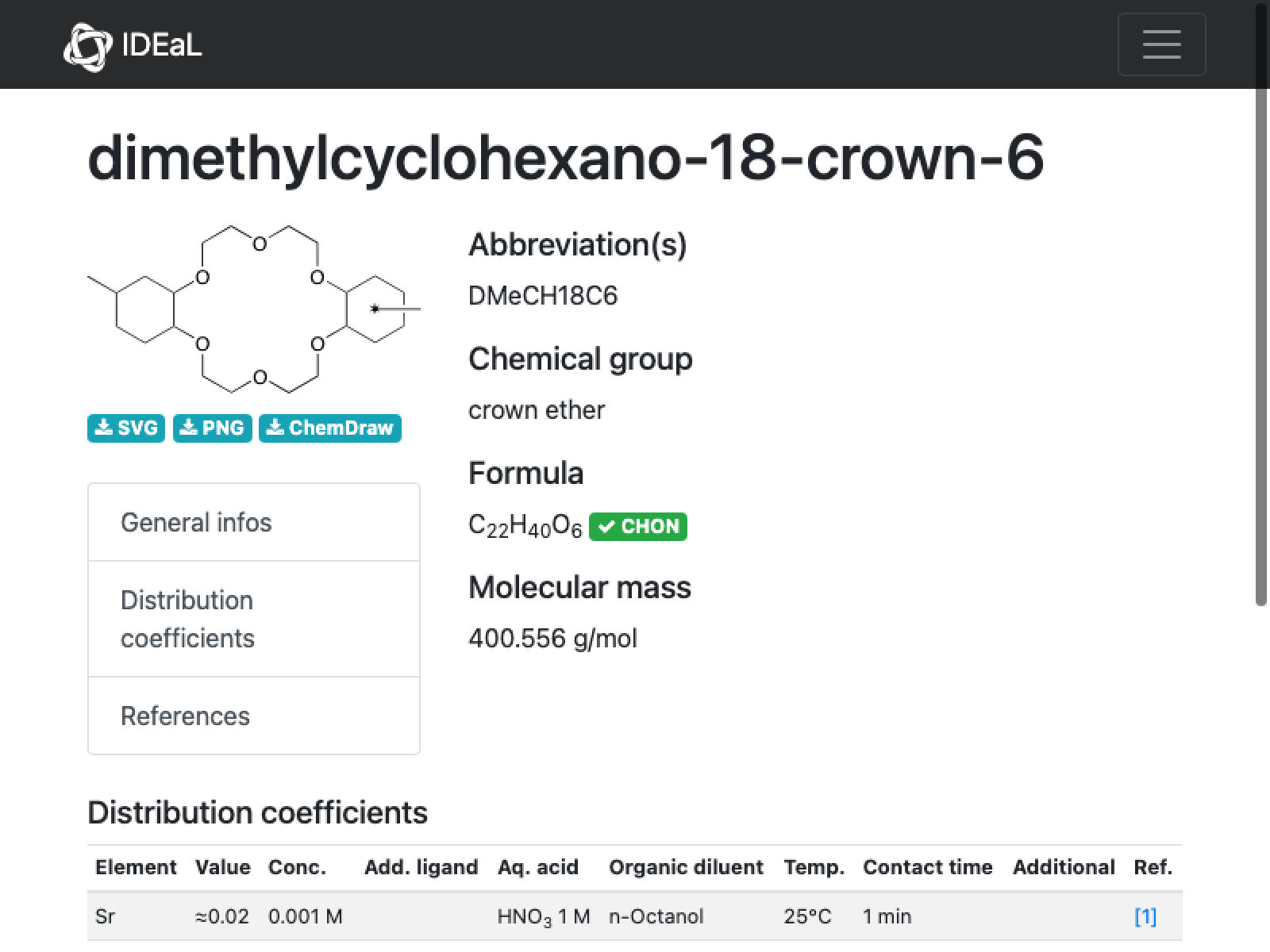
Comment: Looks like the issue here is that the position of the methyl group on the right is not explicit . The cited paper clarifies it to be the 4,4’(5’) compound (purchased commercially), so I will edit it appropriately, starting from downloading the CDX file
outputData[missing[[8]], Molecule["CC1CC2OCCOCCOC3C(CC(C)CC3)OCCOCCOC2CC1"]]
AppendTo[correct, %];

9
missing[[9]]

Same thing as #8, but the isobutyl compound (same source paper)
outputData[missing[[9]], Molecule["CC(C)(C)C1CC2OCCOCCOC3C(CC(C(C)(C)C)CC3)OCCOCCOC2CC1"]]
AppendTo[correct, %];

10
missing[[10]]

WebImage[missing[[10, "url"]]]
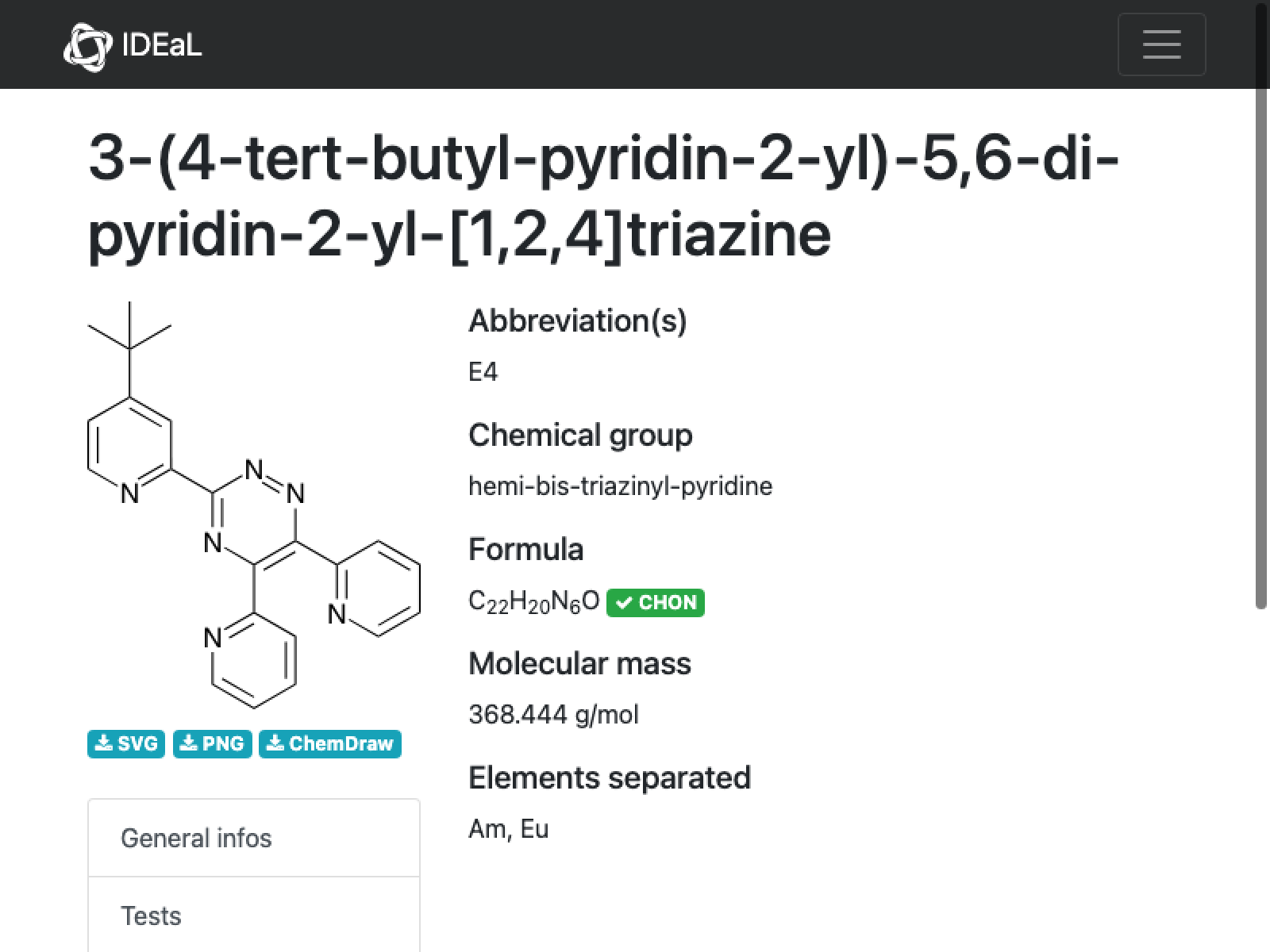
This appears to be a clear case where the formula is wrong–there is not oxygen in this molecule (based on the name); mass is correct
AppendTo[correct, missing[[10]]];
11
missing[[11]]

WebImage@missing[[11, "url"]]
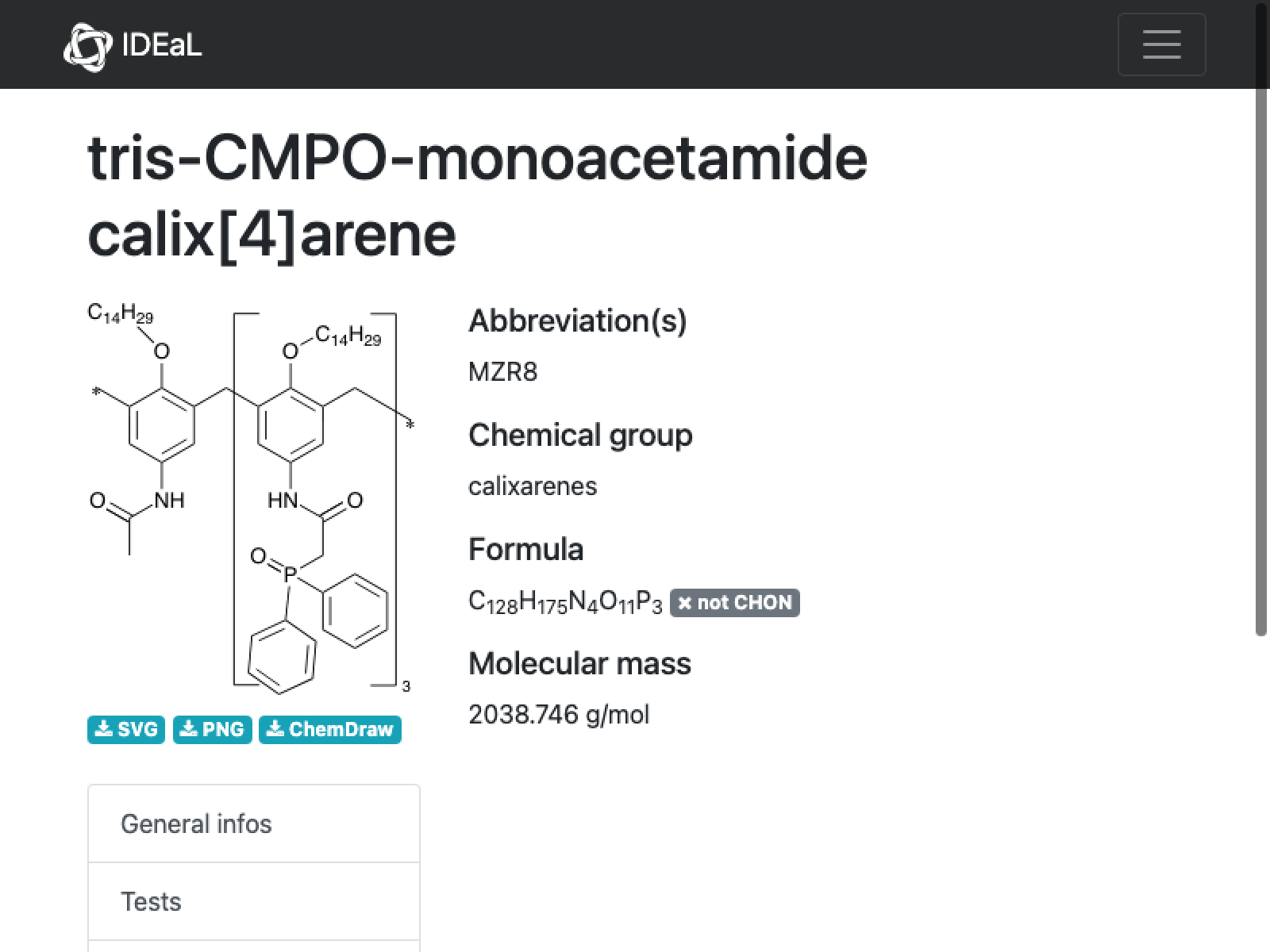
Another ring…best fixed manually in ChemDraw
CopyToClipboard@missing[[11, "SMILES"]]
outputData[missing[[11]], Molecule["O=C(CP(c1ccccc1)(c2ccccc2)=O)Nc3cc(Cc4c(OCCCCCCCCCCCCCC)c(Cc5c(OCCCCCCCCCCCCCC)c(Cc6c(OCCCCCCCCCCCCCC)c(C7)cc(NC(C)=O)c6)cc(NC(CP(c8ccccc8)(c9ccccc9)=O)=O)c5)cc(NC(CP(c%10ccccc%10)(c%11ccccc%11)=O)=O)c4)c(OCCCCCCCCCCCCCC)c7c3"]]
AppendTo[correct, %];

12
missing[[12]]
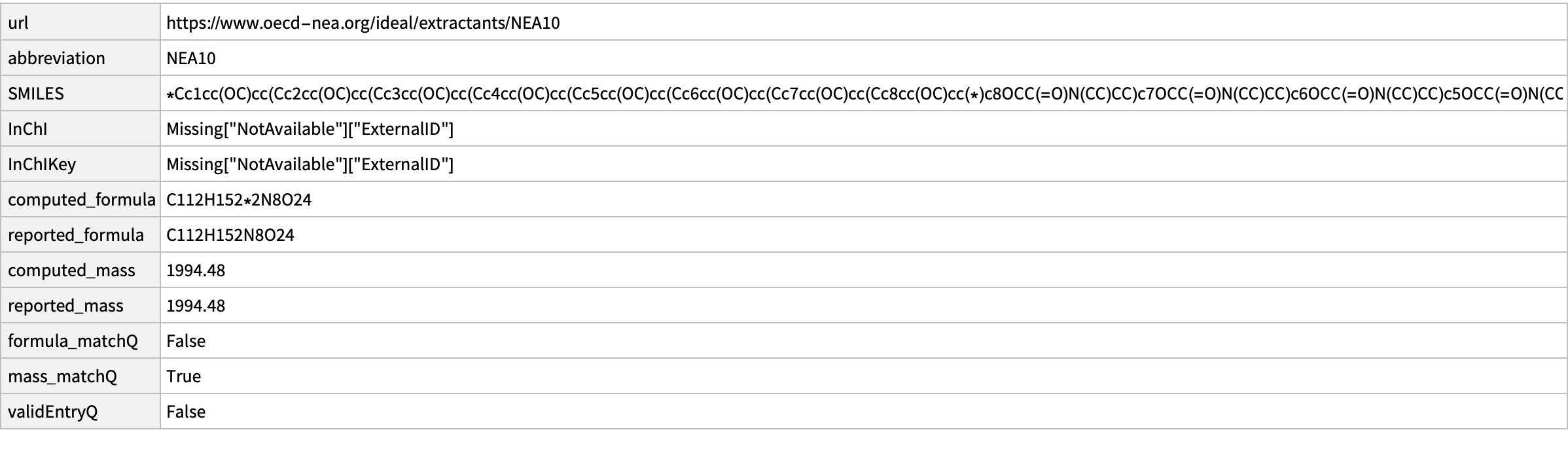
Yet another ring…we know what to do…
CopyToClipboard@missing[[12, "SMILES"]]
outputData[missing[[12]], Molecule["COc1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(C8)cc(OC)c7)cc(OC)c6)cc(OC)c5)cc(OC)c4)cc(OC)c3)cc(OC)c2)c(OCC(N(CC)CC)=O)c(Cc9c(OCC(N(CC)CC)=O)c8cc(OC)c9)c1"]]
AppendTo[correct, %];

13
missing[[13]]

Same story here…
CopyToClipboard@missing[[13, "SMILES"]]
outputData[missing[[13]], Molecule["CCCCCOc1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(C8)cc(OCCCCC)c7)cc(OCCCCC)c6)cc(OCCCCC)c5)cc(OCCCCC)c4)cc(OCCCCC)c3)cc(OCCCCC)c2)c(OCC(N(CC)CC)=O)c(Cc9c(OCC(N(CC)CC)=O)c8cc(OCCCCC)c9)c1"]]
AppendTo[correct, %];

14
missing[[14]]

Seems to be a common story for all the NEA* ligands…
CopyToClipboard@missing[[14, "SMILES"]]
outputData[missing[[14]], Molecule["CC(C)(C)c1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(C8)cc(C(C)(C)C)c7)cc(C(C)(C)C)c6)cc(C(C)(C)C)c5)cc(C(C)(C)C)c4)cc(C(C)(C)C)c3)cc(C(C)(C)C)c2)c(OCC(N(CC)CC)=O)c(Cc9c(OCC(N(CC)CC)=O)c8cc(C(C)(C)C)c9)c1"]]
AppendTo[correct, %];

15
missing[[15]]
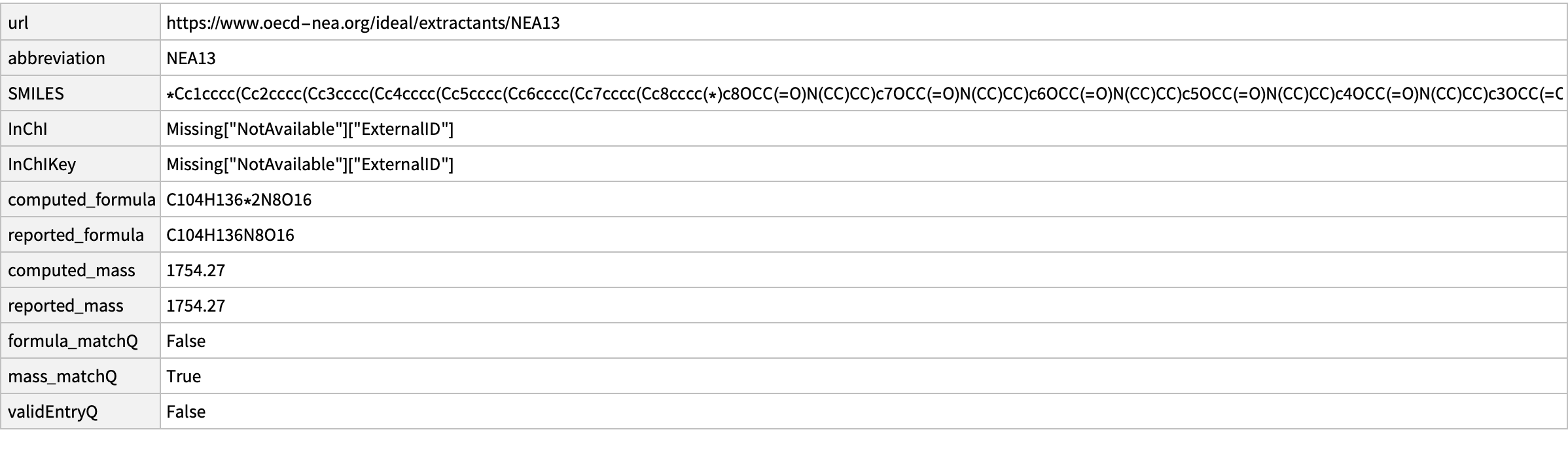
CopyToClipboard@missing[[15, "SMILES"]]
outputData[missing[[15]], Molecule["O=C(N(CC)CC)COc1c2cccc1Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(Cc8c(OCC(N(CC)CC)=O)c(Cc9c(OCC(N(CC)CC)=O)c(C2)ccc9)ccc8)ccc7)ccc6)ccc5)ccc4)ccc3"]]
AppendTo[correct, %];

16
missing[[16]]
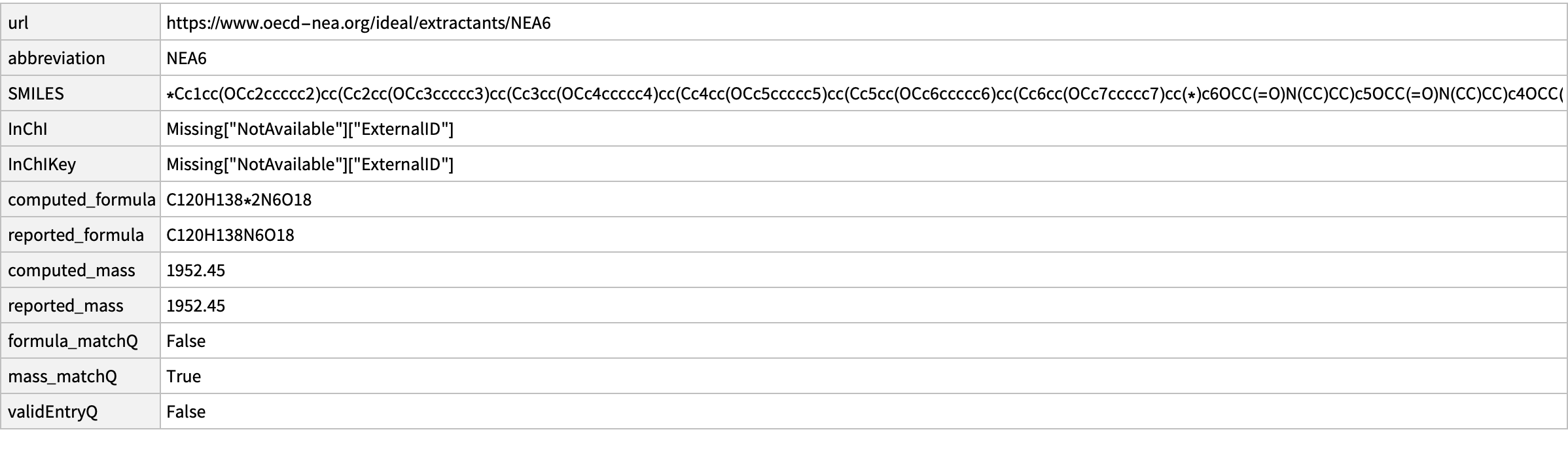
CopyToClipboard@missing[[16, "SMILES"]]
outputData[missing[[16]], Molecule["O=C(N(CC)CC)COc1c2cc(OCc3ccccc3)cc1Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(Cc8c(OCC(N(CC)CC)=O)c(C2)cc(OCc9ccccc9)c8)cc(OCc%10ccccc%10)c7)cc(OCc%11ccccc%11)c6)cc(OCc%12ccccc%12)c5)cc(OCc%13ccccc%13)c4"]]
AppendTo[correct, %];

17
missing[[17]]

CopyToClipboard@missing[[17, "SMILES"]]
outputData[missing[[17]], Molecule["CCCCCCCCOc1cc(Cc2c(OCC(N(CC)CC)=O)c(Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(C6)cc(OCCCCCCCC)c5)cc(OCCCCCCCC)c4)cc(OCCCCCCCC)c3)cc(OCCCCCCCC)c2)c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c6cc(OCCCCCCCC)c7)c1"]]
AppendTo[correct, %];

18
missing[[18]]

CopyToClipboard@missing[[18, "SMILES"]]
outputData[missing[[18]], Molecule["O=C(N(CC)CC)COc1c2cccc1Cc3c(OCC(N(CC)CC)=O)c(Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(C2)ccc7)ccc6)ccc5)ccc4)ccc3"]]
AppendTo[correct, %];

19
missing[[19]]
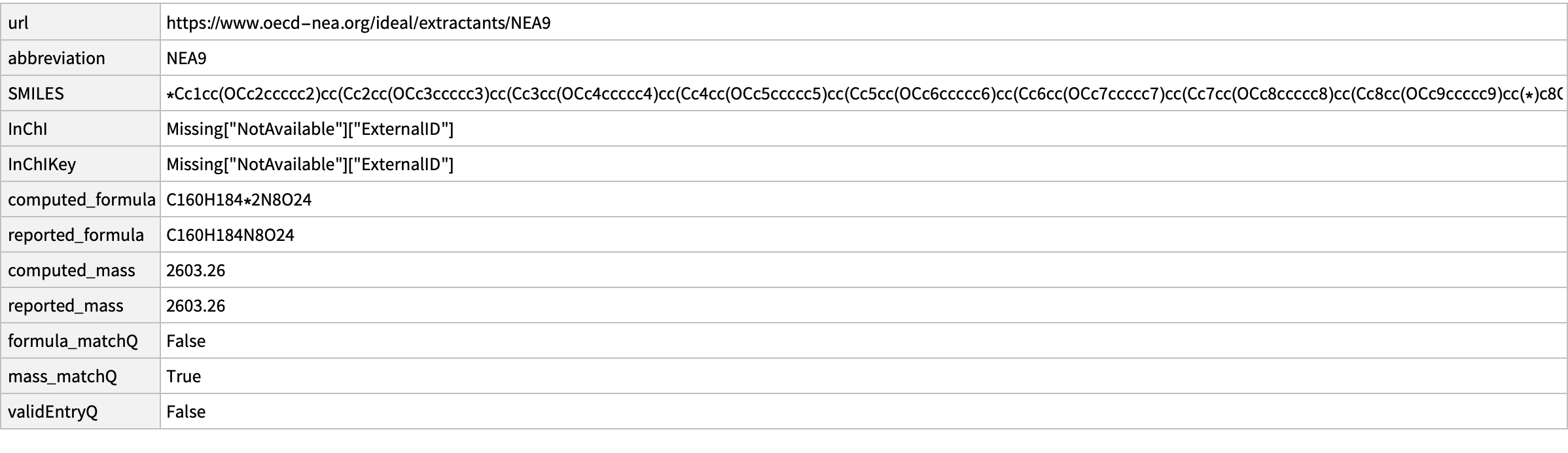
CopyToClipboard@missing[[19, "SMILES"]]
outputData[missing[[19]], Molecule["O=C(N(CC)CC)COc1c2cc(OCc3ccccc3)cc1Cc4c(OCC(N(CC)CC)=O)c(Cc5c(OCC(N(CC)CC)=O)c(Cc6c(OCC(N(CC)CC)=O)c(Cc7c(OCC(N(CC)CC)=O)c(Cc8c(OCC(N(CC)CC)=O)c(Cc9c(OCC(N(CC)CC)=O)c(Cc%10c(OCC(N(CC)CC)=O)c(C2)cc(OCc%11ccccc%11)c%10)cc(OCc%12ccccc%12)c9)cc(OCc%13ccccc%13)c8)cc(OCc%14ccccc%14)c7)cc(OCc%15ccccc%15)c6)cc(OCc%16ccccc%16)c5)cc(OCc%17ccccc%17)c4"]]
AppendTo[correct, %];

20
missing[[20]]

URL[missing[[20, "url"]]]

Comment: Computed and reported formulas are the same and seem consistent with the name. The mass seems low by 3 AMU (?)–maybe just a typo. In any case, it does not matter much, as there are no distribution coefficients reported.
AppendTo[correct, missing[[20]]];
21
missing[[21]]

WebImage[missing[[21, "url"]]]
URL@missing[[21, "url"]]

This is an interesting case as there are few problems: (i). The listed formula appears to be wrong (too many hydrogens); (ii) The LLM-scraped formula is wrong (apparently OpenAI did not like the dot, or at least determined that as an end…a reasonable mistake); (iii) The InChI representation correctly capture the idea that this is a salt, whereas the SMILES does not (?)
Map[MoleculePlot@Molecule[#] &]@{ missing[[21, "InChI"]], missing[[21, "SMILES"]]} // GraphicsRow
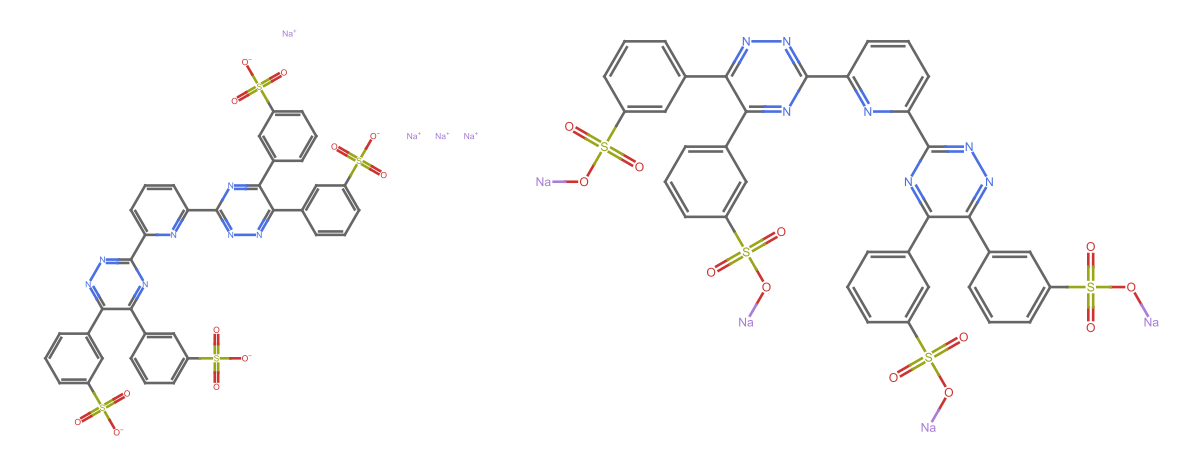
We can fix the SMILES by round tripping it from an initial InChI
MoleculePlot@Molecule@#["CanonicalSMILES"] &@Molecule@missing[[21, "InChI"]]
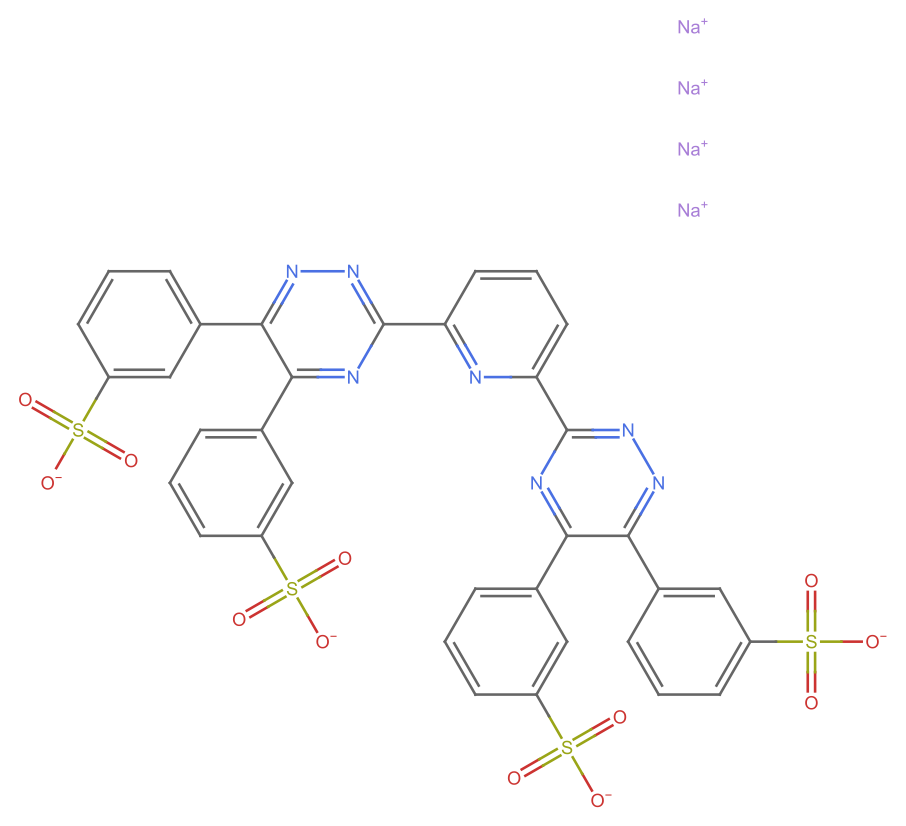
outputData[missing[[21]], Molecule@missing[[21, "InChI"]]]
AppendTo[correct, %];

22
missing[[22]]
URL[%["url"]]
WebImage[%]


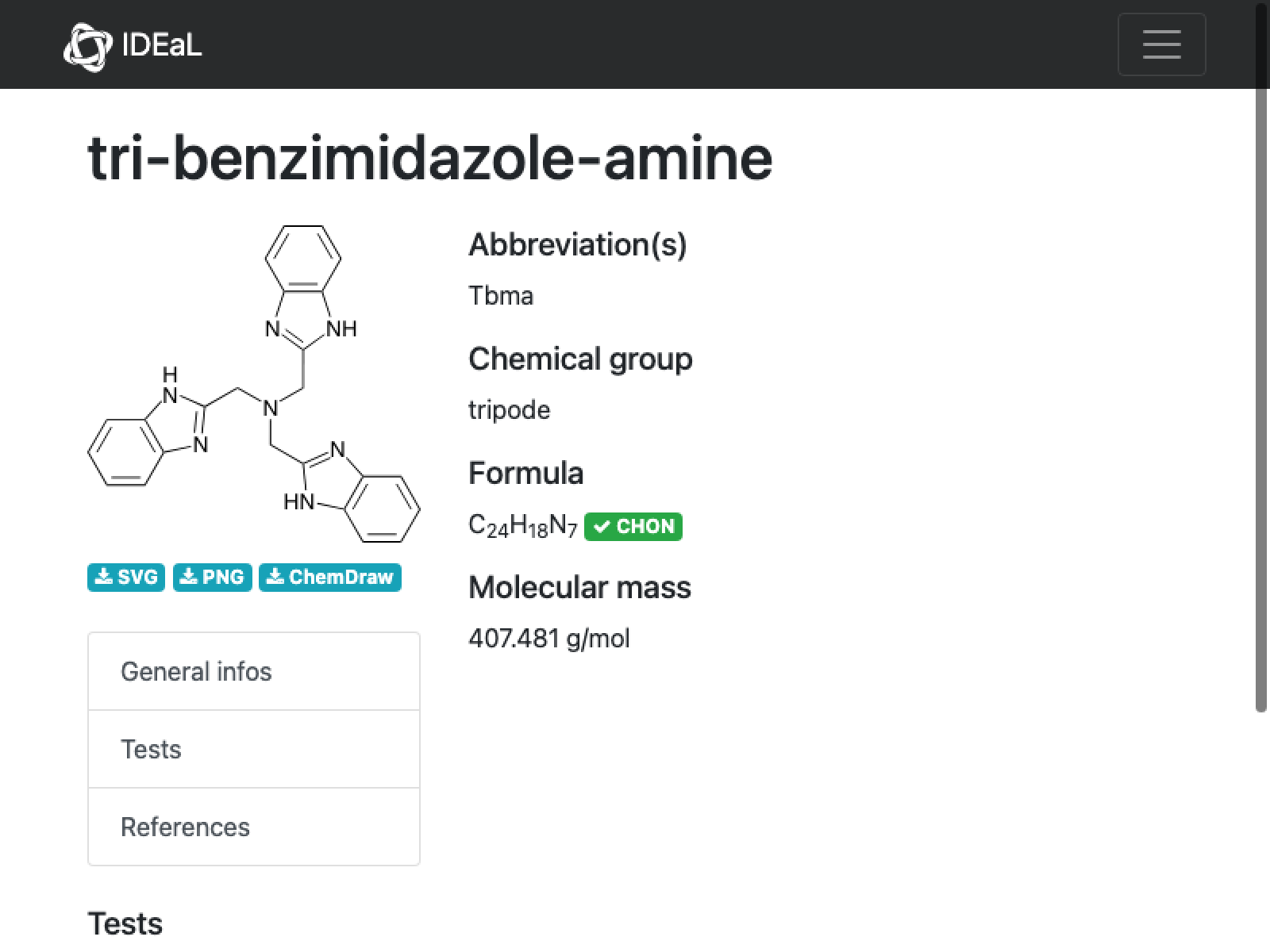
Comment: Hmmm….no tests performed. Mol weight seems fine, which suggests the formula is goofy. But in the end it will not matter much
AppendTo[correct, missing[[22]]];
23
missing[[23]]
URL[%["url"]]
WebImage[%]


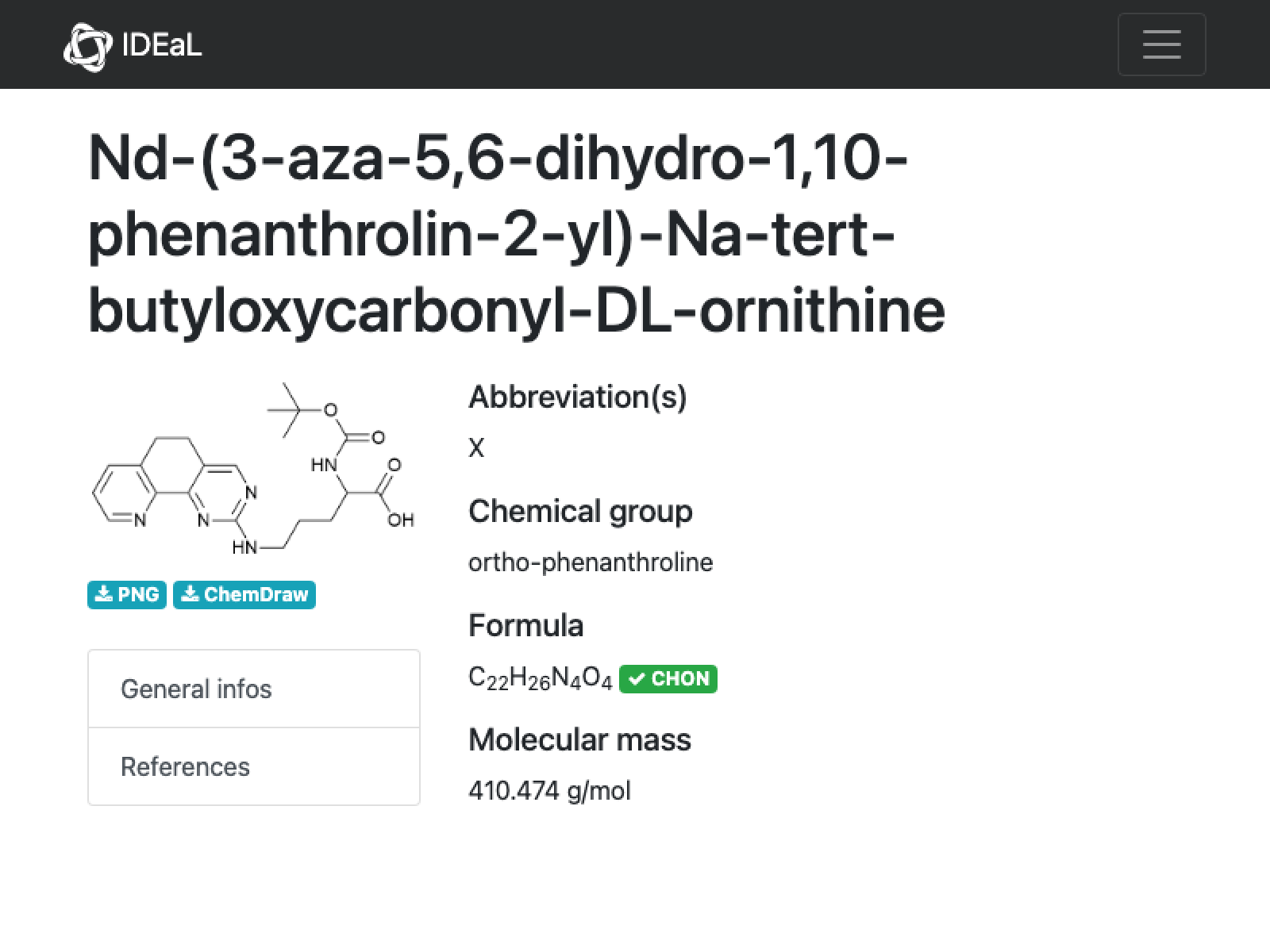
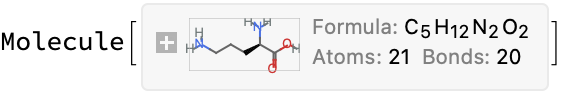
Not much to go on here. The structure backbone indeed looks like an ornithine (note that this is likely a racemate given the name) and the other parts seem to match. There should totally be 5 nitrogens in this (3 from the aza-phenanthroline plus two from the ornithine), so this looks like a bad formula and bad mass on IDEaL. Not that it matters much, because there is no data reported…
Molecule["ornithine"]
Molecule["3-aza-5,6-dihydro-1,10-phenanthroline"]
AppendTo[correct, missing[[23]]];
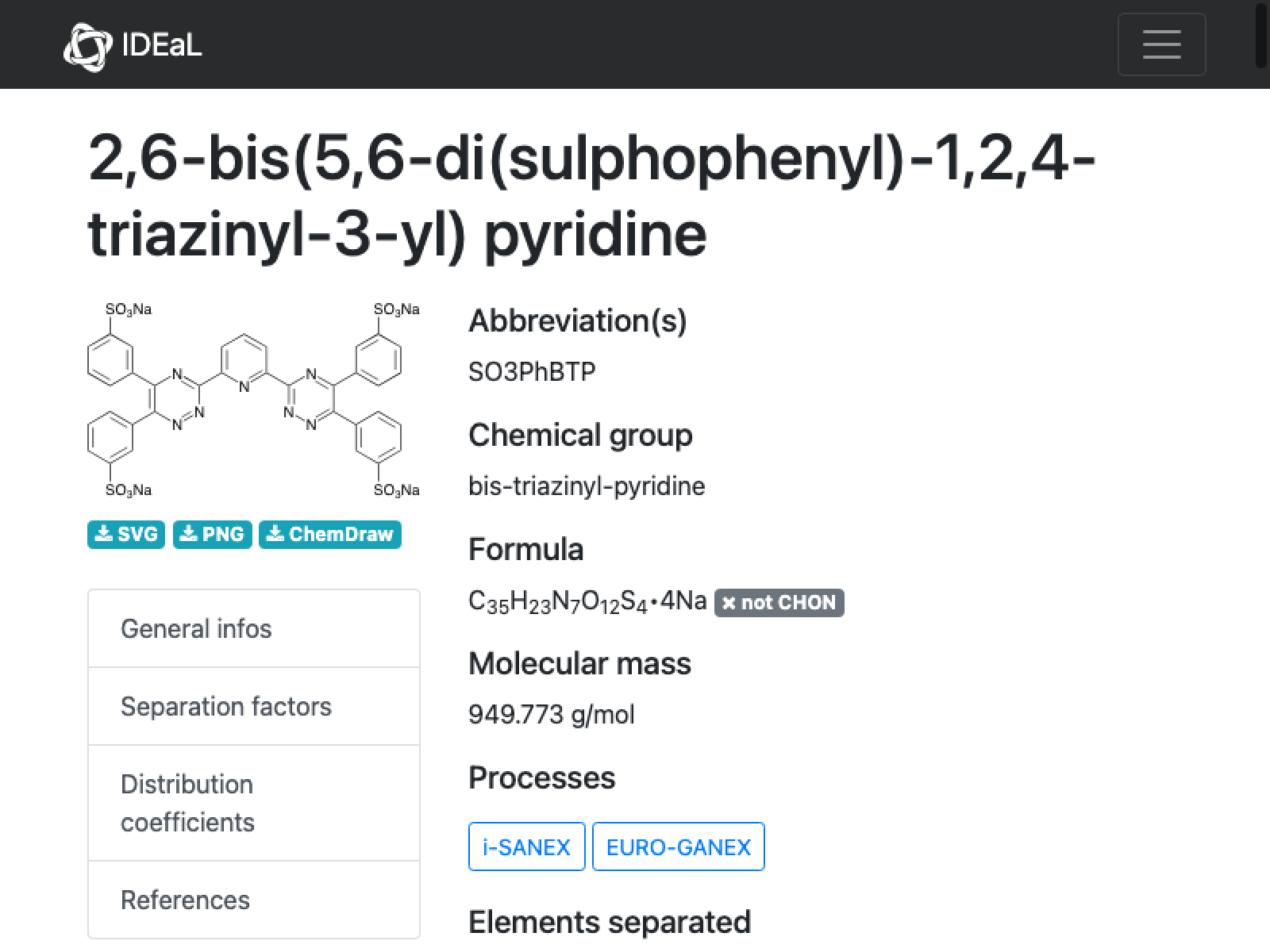
24
missing[[24]]
URL[%["url"]]
WebImage[%]


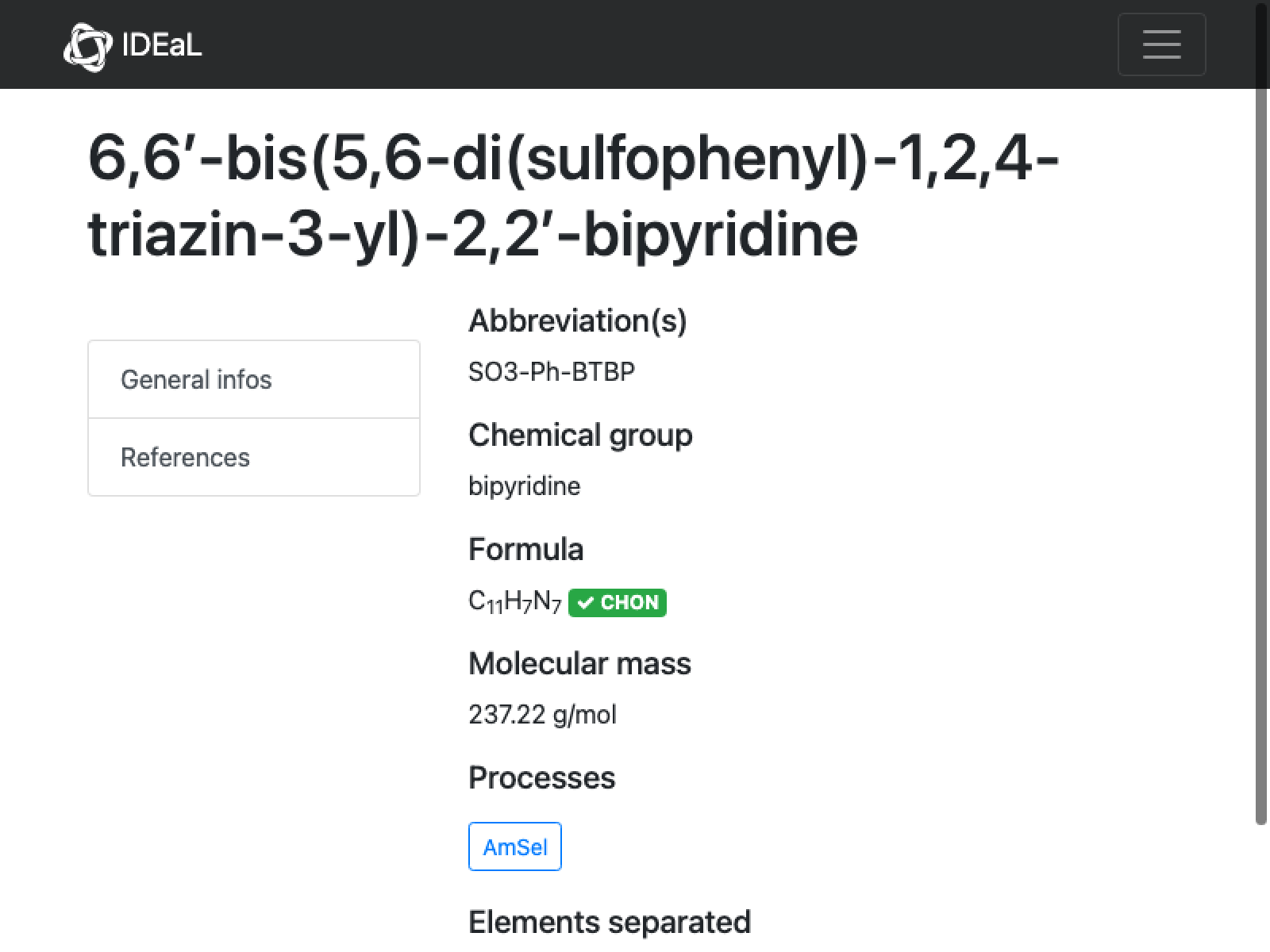
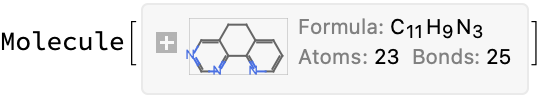
There is not much to go on here; apparently we rejected it initially because the name is not unambiguous (placement of the sulfates on the phenyl):
mol = Molecule["6,6'-bis(5,6-di(sulfophenyl)-1,2,4-triazin-3-yl)-2,2'-bipyridine"]
MoleculePlot[%]
MoleculeValue[%%, {"MolecularFormulaString", "MolarMass"}]
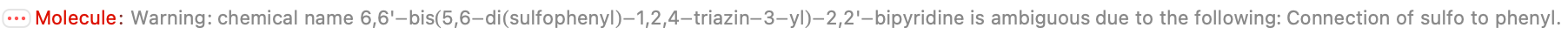

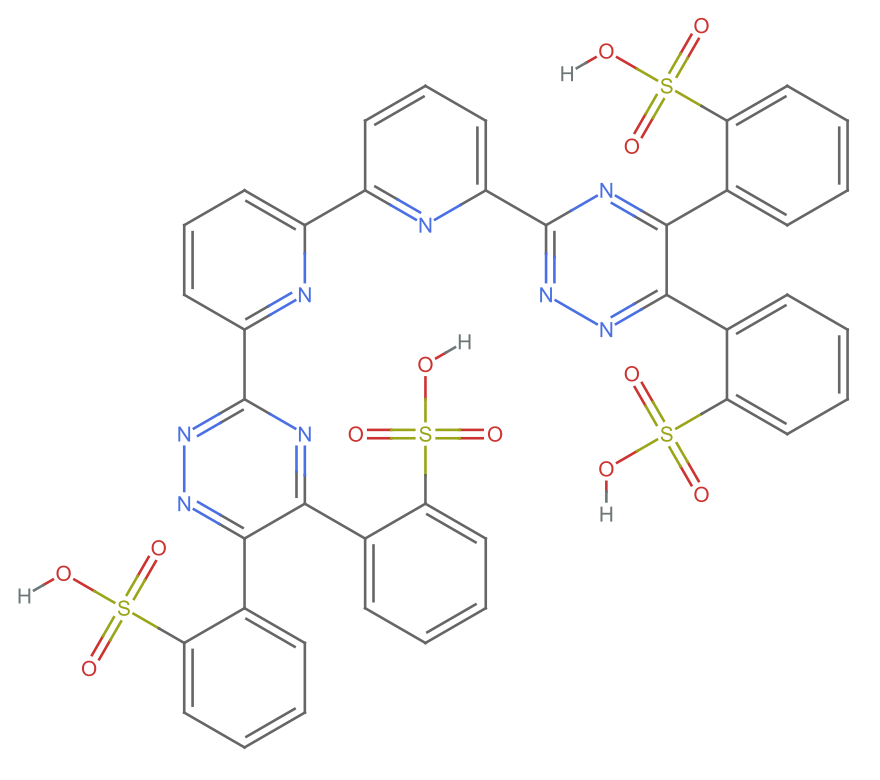
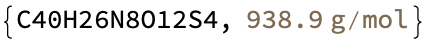
This is totally not the same molecular formula and mass given on the IDEaL website. However, this structure looks consistent with the stated name–I do not see how you would get the bis di sulfulophenyl with fewer atoms. And there is no other data, so my decision is to run with this.
outputData[missing[[24]], Molecule[mol]]
AppendTo[correct, %];

25
missing[[25]]
URL[%["url"]]
WebImage[%]


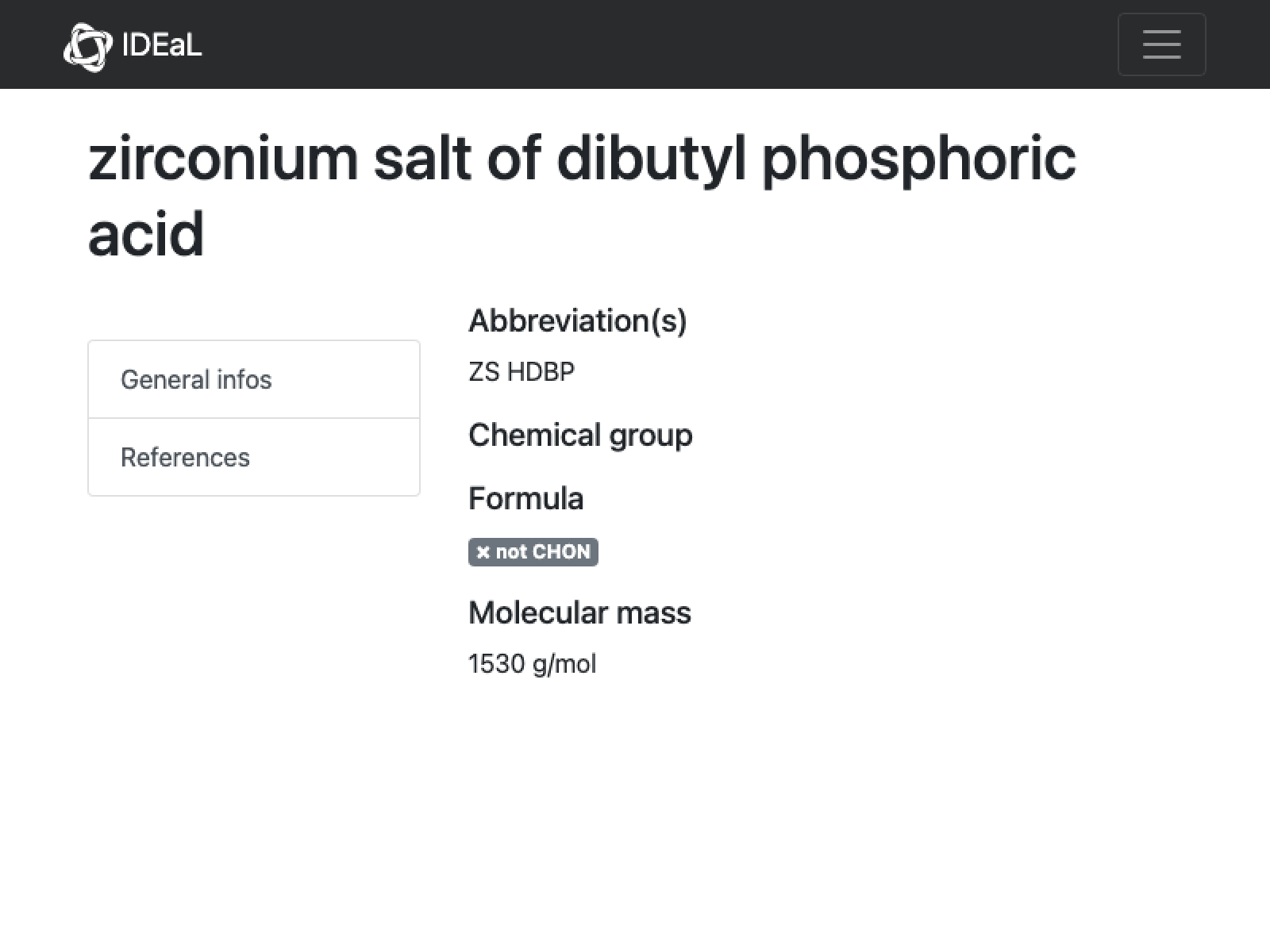
Once again, not much to go on here. I found a paper which uses this for a separation with ratios of HDBP:Zr of 9, but that makes the mass too high; the closest I can get to the stated mass is 7:1 and even then it is not a perfect match:
Molecule["dibutyl phosphoric acid"]
%["MolarMass"]*7 + ElementData["Zr", "MolarMass"]
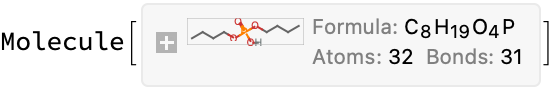
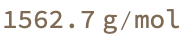
Modest proposal: It does not matter much, because there are no references. So I will just put in a 1:1 stoichiometry and call it a day
mol = Molecule@StringJoin[
"[Zr].",
Molecule["dibutyl phosphoric acid"]["CanonicalSMILES"]]
MoleculePlot[%]
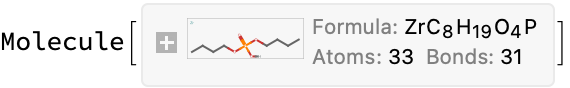
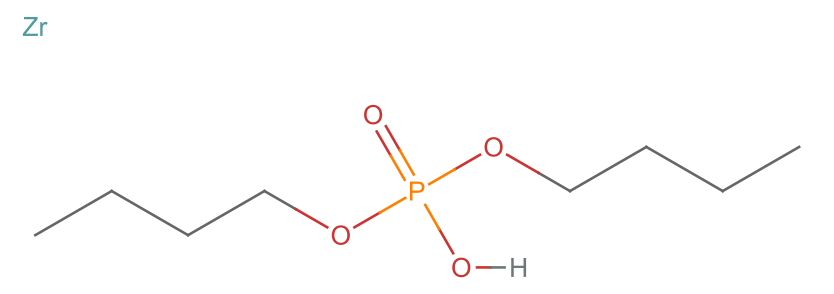
outputData[missing[[25]], mol]
AppendTo[correct, %];

Conclusion
Did we get them all?
(Length[excluded] + Length[correct] ) == Length[missing]
(*True*)
Remember, we excluded one case:
excluded[[1]]

Merge these new results with the correct values found in the last episode; the resulting file can be downloaded here.
With[
{previous = Import["2023.10.16_all_correct_entries.xlsx", {"Dataset", 1}, "HeaderLines" -> 1]},
Join[previous, correct]];
Export["2023.10.17_all_correct_entries.xlsx", %];
Length[%%] (*count how many we have*)
(*438*)
ToJekyll["Parsing Molecular Identifiers From the Ideal Database, part 3", "mathematica chemdraw science"]