Parsing molecular identifiers from the IDEaL Database
[llm mathematica science gpt3.5 Suppose you (not-so-hypothetically) want to screen-scrape the IDEaL database in order to build your own f-element separation database. While it is straightforward to task a student with scraping the data tables, extracting machine-readable chemical structures is more challenging, as they do not provide standard chemical identifiers such as SMILES, InChI, etc. However, they do provide Chemdraw files and IUPAC-ish names which we can try to parse. This also provides an opportunity to do some LLM-based screen scraping. (Spoiler alert: We uncover approximately 105/438 entries with unresolvable structures, including 6 cases where the stated molecular formula is inconsistent with the stated molecular mass.) Mathematica to the rescue…
Get a list of all the links to each compound
Start by getting a list of all the ligand URLs:
homepage = "https://www.oecd-nea.org/ideal/";
links = URLBuild /@ URLParse /@ Select[StringContainsQ["/extractants/"]]@Import[homepage, "Hyperlinks"];
Length[%]
(*439*)
Why round-trip the retrieved Hyperlinks through URLBuild and URLParse? Hyperlink importing returns an unescaped string (i.e., replaces %20 with a blankspace). But we need these to actually retrieve the files! If you do not do this, then the following hardExample will throw an error when we try to retrieve files:
hardExample = links[[50]]
(*"https://www.oecd-nea.org/ideal/extractants/AFR%20Nitro"*)
Comment: I did not know this was a problem when I started, and only appreciated it when I started applying the functions to the bulk job.
Retrieve data for each compound
In this section we will run through the various extraction functions. To test them we will try the following demonstration example:
example = "https://www.oecd-nea.org/ideal/extractants/CB4";
Abbreviation
The compound abbreviation names are the last part of the URL path. We use URLParse to remove the %-escaping characters (which we added in the round-tripping function above):
moleculeAbbreviation[url_String] := Last@Lookup["Path"]@URLParse[url]
(*demo*)
moleculeAbbreviation[example]
moleculeAbbreviation[hardExample]
(*"CB4"*)
(*"AFR Nitro"*)
Retrieve and parse the Chemdraw file
Yeah, Mathematica can Import Chemdraw .cdx files. Again, the extracted Hyperlinks get unescaped, so we roundtrip the retrieved URL through URLBuild/URLParse to make it readable:
(*given an extractant URL return the Molecule found in the linked Chemdraw file*)
moleculeFromChemdraw[url_] := With[
{cdxFileURL = SelectFirst[StringContainsQ[".cdx"]]@Import[url, "Hyperlinks"]},
Import@URLBuild@URLParse[cdxFileURL]
]
(*demo*)
exampleMol = moleculeFromChemdraw[example]
moleculeFromChemdraw[hardExample]
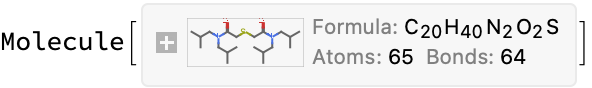
Extract other information using GPT-3.5
Each HTML file contains some preliminary information (untagged) about the molecule name, molecular formula and molecular mass:
text = Import[example, "Plaintext"]
StringLength[text]
(*"IDEaL Browse Search Back-office 2,2'-thiobis(N,N-diisobutylacetamide) SVG PNG ChemDraw General infos Tests References Abbreviation(s) CB4 Chemical group thiodiglycolamide Formula C 20 H 40 N 2 O 2 S not CHON Molecular mass 372.612 g/mol Elements separated Eu, Pu, Am, Cm Tests Test carried out Test type Conditions Ref.Extraction [1] References CEA"*)
(*460*)
We could try to write a parser for this, but it seems tedious, so instead I decided to use an LLMFunction to extract the information; we will trim the plaintext down to a few thousand characters to make sure it fits into the context window. In practice, the information we care about is near the top, in the first 500-1000 characters. (I had originally experimented with three separate functions, but ultimately combined them so that I can make a single API call and thus save a bit of time and a few pesos. However, as we shall see below, it is also useful to break these out to deal with certain debugging tasks.) As of this writing, LLMFunction defaults is to use OpenAI GPT-3.5 with temperature = 0. The progress reports are annoying when you apply this to a batch of results, so we turn it off with an undocumented ProgressReporting option:
llmExtractor = LLMFunction["What is the compound name, molecular formula, and molecular mass in the following text? Return only the name, formula and number with no units or other information. Return the results as a JSON dictionary with keys compound_name, molecular_formula, and molecular_mass.\n``",
ProgressReporting -> False
];
compoundInformation[url_] := With[
{text = StringTake[#, UpTo[1500]] &@Import[url, "Plaintext"]},
ImportString[#, "RawJSON"] &@llmExtractor[text]
]
(*demo*)
exampleResult = compoundInformation[example]
(*<|"compound_name" -> "2,2'-thiobis(N,N-diisobutylacetamide)", "molecular_formula" -> "C20H40N2O2S", "molecular_mass" -> "372.612"|>*)
Comment: I am actually pleasantly surprised at how well this worked. If you look at the string, it is quite ugly and I suspect there are many edge cases for a simple regexp. What I like about this use case is that it fails in a transparent way—if we get bad data in our consistency check then we will just have to manually correct the data.
Addendum (06 Nov 2023): The new function calling API options may allow you to simplify the prompt for returning JSON, in case you are trying this in the future.
We can check for consistency against the listed molecular formula and molecular mass (rounding the latter to the nearest integer as good-enough. In practice, getting the molecular formula correct should also get the mass correct, but I like a belt-and-suspenders approach.
moleculeConsistentQ[mol_Molecule, info_Association] := With[
{formula = MoleculeProperty["MolecularFormulaString"]@mol,
mass = QuantityMagnitude@ MoleculeProperty["MolarMass"]@mol},
(formula == info["molecular_formula"]) && (Round[mass] == Round[ Interpreter["Number"][info["molecular_mass"] ]])
]
(*demo*)
moleculeConsistentQ[exampleMol, exampleResult]
(*True*)
We also want to have backup ways of trying to assign the molecules, if some of these are messed up. We can try to parse the IUPAC name or kick it to PubChem. Both of these might fail, so we use the Enclose[Confirm[]] pattern, and just return empty molecules if it fails. I overload the function definitions to take both strings and the information dictionary returned from the LLM interpretation just for convenience:
interpretName[name_String] := Enclose[
ConfirmQuiet[Molecule[name]],
Molecule[""] &]
interpretName[info_Association] := interpretName@info["compound_name"]
(*demo*)
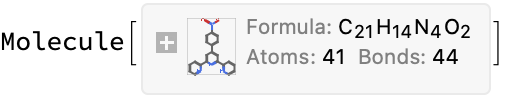
interpretName[exampleResult]
moleculeConsistentQ[%, exampleResult]
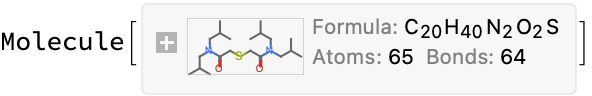
(*True*)
Similarly use the built-in Pubchem service to do a synonym lookup; again, if we get a warning message about the compound not being present, then we just return an empty molecule. In this case, this is not actually a molecule in PubChem, so this shows the values of different methods:
interpretNameWithPubchem[name_String] := With[
{pubchemResults = Enclose[
ConfirmQuiet[
ServiceExecute["PubChem", "CompoundProperties", {"Name" -> name, "Property" -> "InChI"}]],
{<|"InChI" -> ""|>} &]},
Molecule@First@Lookup[#, "InChI", ""] &@Normal@pubchemResults]
interpretNameWithPubchem[info_Association] := interpretNameWithPubchem@info["compound_name"]
(*demo*)
interpretNameWithPubchem[exampleResult]
moleculeConsistentQ[%, exampleResult]

(*False*)
Note that IDEaL also has images of the molecules, and one could use MoleculeRecognize to parse them. However, in some isolated tests, I found that these were pretty bad (because of transparencies, abbreviations, etc. in the images), so it is not worth implementing.
Putting it all together:
Now that we have defined all the pieces, we can combine them to generate the output:
Define the function
We process each link one at a time, defining the output for each line as an Association. If the molecule is FUBAR, then we just report it as missing. Finally we try reading the molecule from the chemdraw file, the name, and pubchem (in order) until one of them passes the consistency test (w.r.t. molecular formula and molecular weight):
(*we have a valid molecule! yay! *)
outputData[url_String, mol_?MoleculeQ] := Association[
"url" -> url,
"abbreviation" -> moleculeAbbreviation[url],
"SMILES" -> MoleculeValue[mol, "CanonicalSMILES"],
"InChI" -> MoleculeValue[mol, "InChI"]["ExternalID"],
"InChIKey" -> MoleculeValue[mol, "InChIKey"]["ExternalID"]
]
(* something is fishy, so fail *)
outputData[url_String, _] := Association[
"url" -> url,
"abbreviation" -> moleculeAbbreviation[url],
"SMILES" -> Missing[],
"InChI" -> Missing[],
"InChIKey" -> Missing[]
]
(*I don't really like this nested if, but it does the job...*)
processURL[url_String] := Module[
{mol = moleculeFromChemdraw[url],
info = compoundInformation[url]},
If[ moleculeConsistentQ[mol, info],
outputData[url, mol], (*if true*)
mol = interpretName[info]; (*else try the name...*)
If[moleculeConsistentQ[mol, info],
outputData[url, mol], (*if true*)
mol = interpretNameWithPubchem[info]; (*else try pubchem...*)
If[moleculeConsistentQ[mol, info],
outputData[url, mol], (*if true*)
outputData[url, Missing[]] (*it failed...*)
]]]]
(*demo*)
processURL[example]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/CB4", "abbreviation" -> "CB4", "SMILES" -> "CC(C)CN(CC(C)C)C(=O)CSCC(=O)N(CC(C)C)CC(C)C", "InChI" -> "InChI=1S/C20H40N2O2S/c1-15(2)9-21(10-16(3)4)19(23)13-25-14-20(24)22(11-17(5)6)12-18(7)8/h15-18H,9-14H2,1-8H3", "InChIKey" -> "DNJRGLMKJFJUJD-UHFFFAOYSA-N"|>*)
Map it over the list of entries
Showtime! Map this over links and convert it to a Dataset. I use a ParallelMap so that I can run multiple LLM calls simultaneously (waiting for the response is the bottleneck). It took less than 5 minutes to run. However some errors came out. We will deal with these later (vide infra):
results = ParallelMap[processURL, links] // Dataset



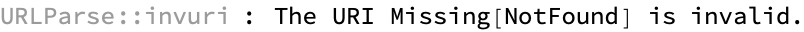
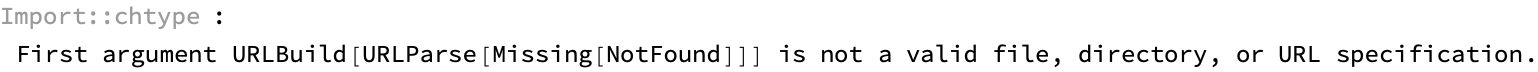
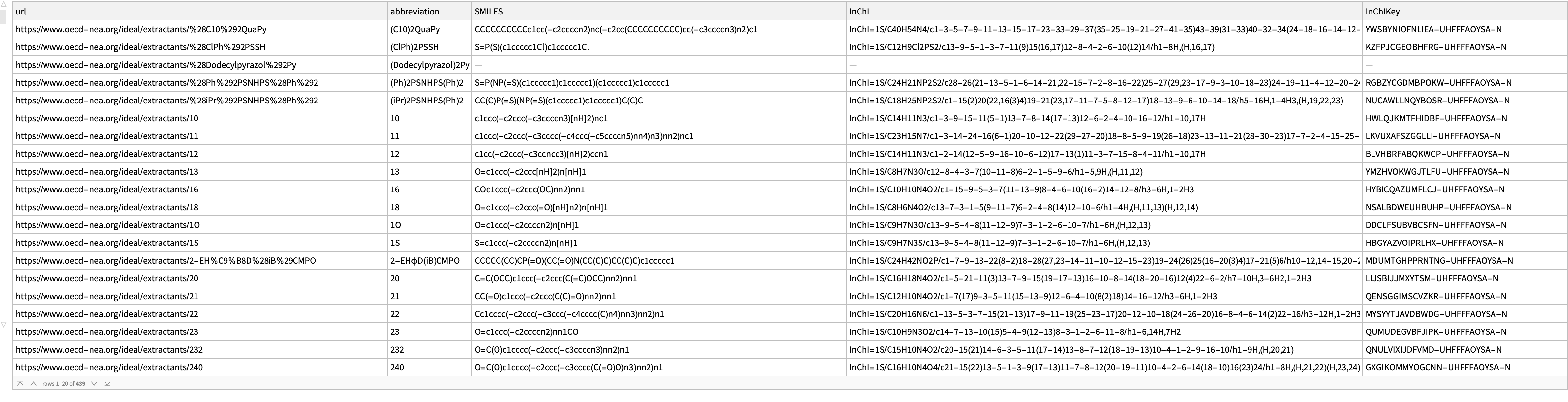
How many unresolved entries are there? Note that these are not computational errors but instead are cases where we were unable to resolve a structure that passed the consistency test:
Length@Select[MissingQ]@Lookup["SMILES"]@Normal@Select[AssociationQ]@results
(*103*)
Comment: Yikes! This seems bad. But it is probably not so bad in practice, because many of the problematic molecules have no distribution coefficient data? (Spoiler alert: We will care about 75 of them…) So I suggest that rather than try to fill all of these in, that one instead just see which entries are missing after doing the final merge.
Before dealing with the (actual) errors, I save the results obtained so far to avoid giving OpenAI another dime.1 To export as a spreadsheet we must remove rows that are not valid Associations (or stated more positively, we Select only rows that are validly structured before exporting:
SetDirectory@NotebookDirectory[];
Export["2023.10.11_ideal_moleculelist.xlsx", Select[AssociationQ]@results];
DumpSave["2023.10.11_ideal_moleculelist.mx", results];
Investigating the Problem Children
There are a limited number of problem cases; it is possible that the relevant servers just timed out or there are strange unicode errors, or there might be other problems. Given that this is a one-time project, and the problems only affect about 1% of the data, I am not so inclined to solve it rigorously, but instead we can take a look:
problems = Cases[outputData[x_, _] :> x] /@ Normal@Select[(! AssociationQ[#]) &]@results // Flatten
Length[%]
(*{"https://www.oecd-nea.org/ideal/extractants/9H", "https://www.oecd-nea.org/ideal/extractants/HONTA", "https://www.oecd-nea.org/ideal/extractants/SO3-Ph-BTBP", "https://www.oecd-nea.org/ideal/extractants/T2EHDGA", "https://www.oecd-nea.org/ideal/extractants/ZS%20HDBP"}*)
(*5*)
We will address each of these individually, creating a place to store the results:
solutions = Table[Association[], {5}];
1) 9H
This looks like a problem with the ChemDraw file–it contains two disconnected fragments, but the first one seems correct (correct
processURL@problems[[1]]
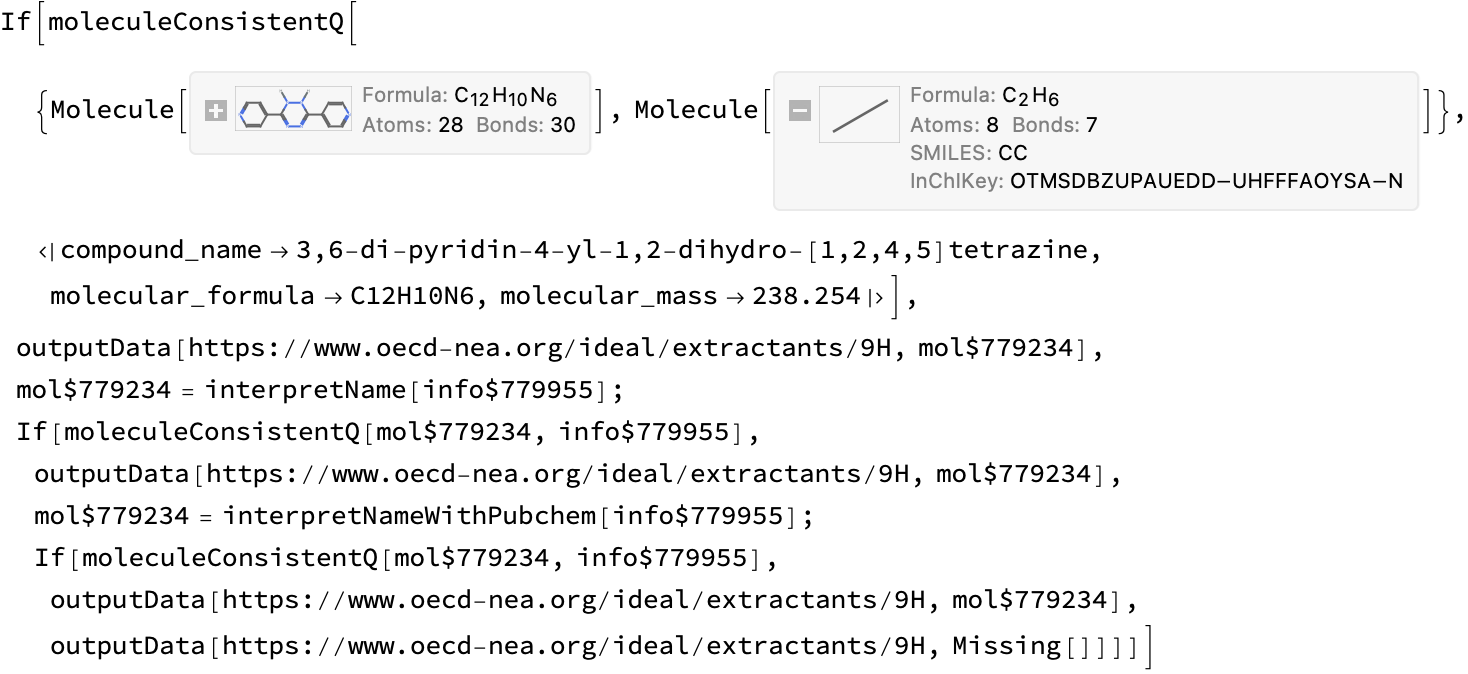
url = problems[[1]];
info = compoundInformation[url]
cdxMol = First@ moleculeFromChemdraw[url]
iupacMol = interpretName[info]
moleculeConsistentQ[iupacMol, info]
solutions[[1]] = outputData[url, iupacMol]
(*<|"compound_name" -> "3,6-di-pyridin-4-yl-1,2-dihydro-[1,2,4,5]tetrazine", "molecular_formula" -> "C12H10N6", "molecular_mass" -> "238.254"|>*)
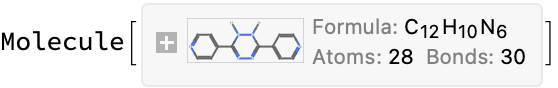
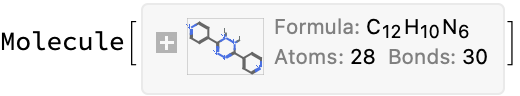
(*True*)
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/9H", "abbreviation" -> "9H", "SMILES" -> "c1cc(C2=NN=C(c3ccncc3)NN2)ccn1","InChI" -> "InChI=1S/C12H10N6/c1-5-13-6-2-9(1)11-15-17-12(18-16-11)10-3-7-14-8-4-10/h1-8H,(H,15,16)(H,17,18)", "InChIKey" -> "QWYDFEKBBFVMQW-UHFFFAOYSA-N"|>*)
2) HONTA
It looks like the problem here is parsing the JSON:
url = problems[[2]];
cdxMol = moleculeFromChemdraw[url]
info = compoundInformation[url]
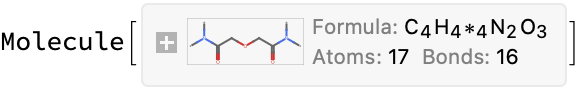


(*$Failed*)
WebImage[url]
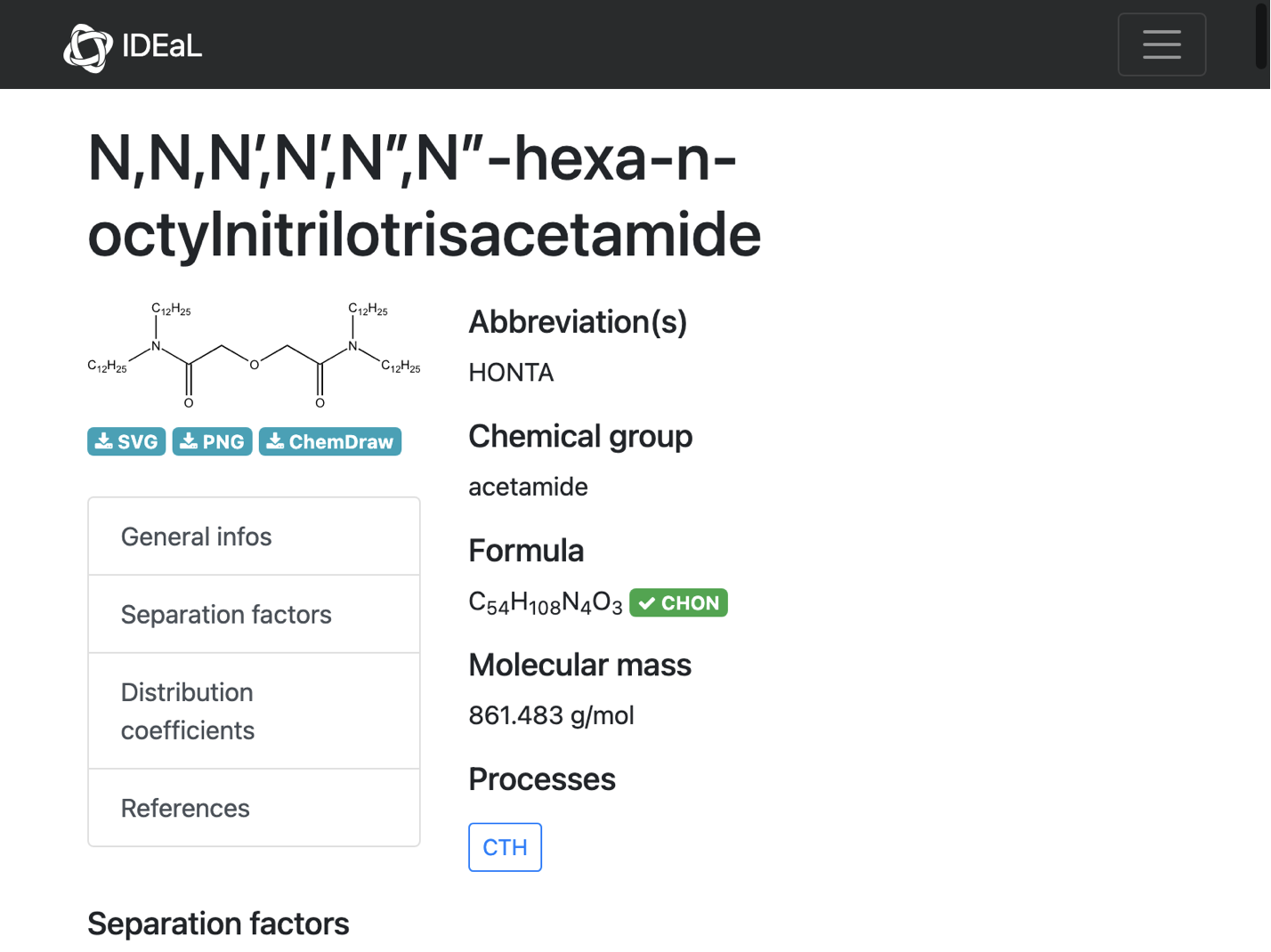
It looks like the problem is with the curly quotes that are being used for indicating primes and double primes (manually copy-pasted below)
iupacMol = Molecule["N,N,N\[CloseCurlyQuote],N\[CloseCurlyQuote],N\[CloseCurlyDoubleQuote],N\[CloseCurlyDoubleQuote]-hexa-n-octylnitrilotrisacetamide"]
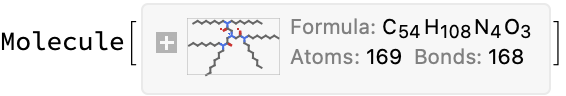
formulaExtract[url_] := LLMFunction["What is the molecular formula in the following text? Return only the formula and no other information.\n ``"]@
StringTake[#, UpTo[1500]] &@Import[url, "Plaintext"]
formulaExtract[url]
(*"C54H108N4O3"*)
solutions[[2]] = outputData[url, iupacMol]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/HONTA", "abbreviation" -> "HONTA", "SMILES" -> "CCCCCCCCN(CCCCCCCC)C(=O)CN(CC(=O)N(CCCCCCCC)CCCCCCCC)CC(=O)N(CCCCCCCC)CCCCCCCC", "InChI" -> "InChI=1S/C54H108N4O3/c1-7-13-19-25-31-37-43-56(44-38-32-26-20-14-8-2)52(59)49-55(50-53(60)57(45-39-33-27-21-15-9-3)46-40-34-28-22-16-10-4)51-54(61)58(47-41-35-29-23-17-11-5)48-42-36-30-24-18-12-6/h7-51H2,1-6H3", "InChIKey" -> "YTTXOOIRNCYALH-UHFFFAOYSA-N"|>*)
3) SO3-Ph-BTBP
url = problems[[3]];
WebImage[url]
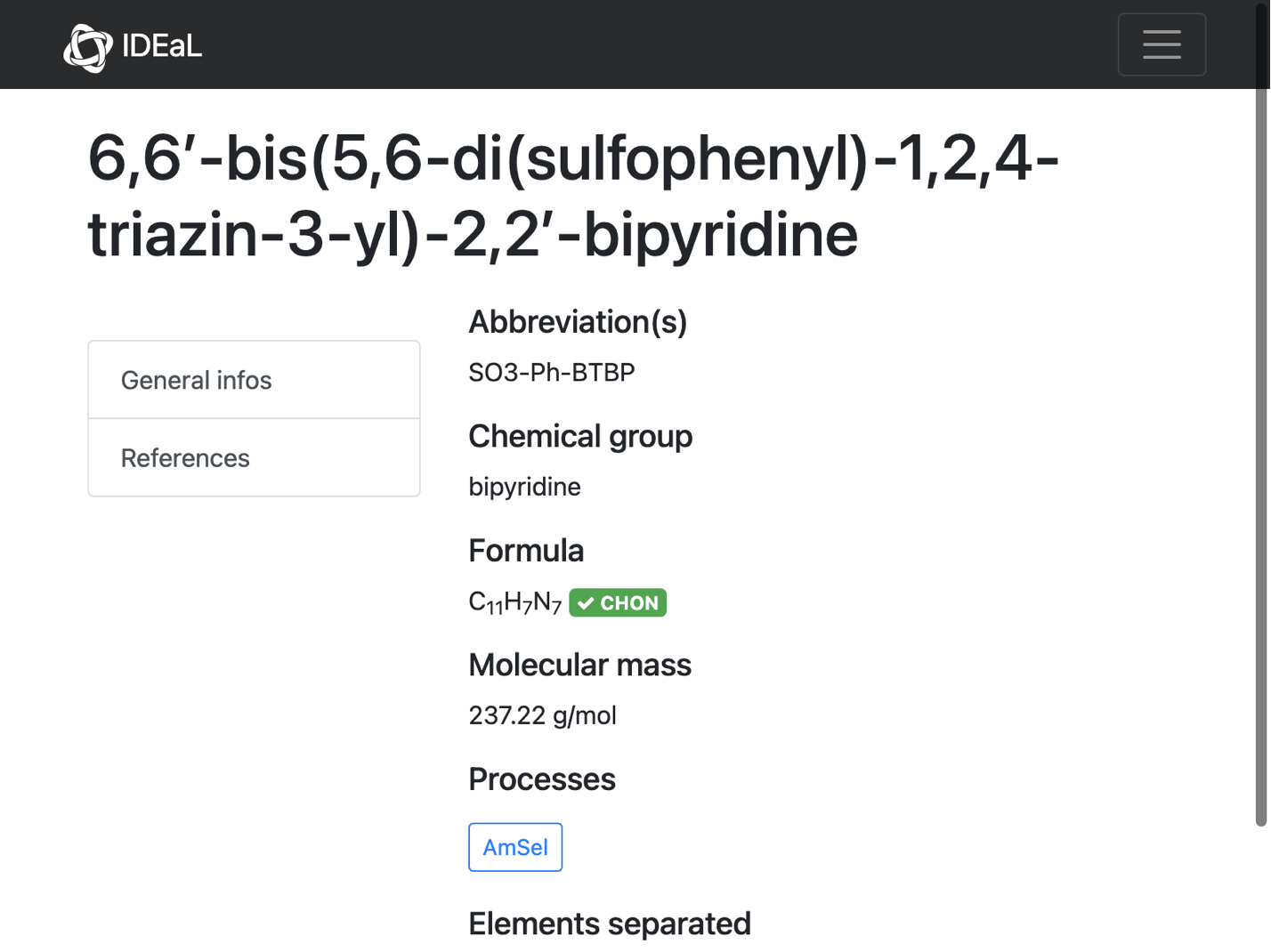
Comment: Looks like the problem here is that there is no chemdraw file AND we have the curly-quote problem
info = compoundInformation[url]


(*$Failed*)
nameExtract[url_] := LLMFunction["What is the compound name in the following text? Return only the name and no other information.\n ``"]@
StringTake[#, UpTo[1500]] &@Import[url, "Plaintext"]
iupacMol = Molecule@nameExtract[url]
formula = formulaExtract[url]
MoleculeValue[iupacMol, "MolecularFormulaString"] == formula


(*"C11H7N7"*)
(*False*)
There appears to be a problem here, but I am not inclined to solve it, as there are no distribution coefficients reported.
solutions[[3]] = outputData[url, Missing[]]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/SO3-Ph-BTBP", "abbreviation" -> "SO3-Ph-BTBP", "SMILES" -> Missing[], "InChI" -> Missing[], "InChIKey" -> Missing[]|>*)
4. T2EHDGA
Another problem with non-unicode characters in the name
url = problems[[4]];
WebImage[url]
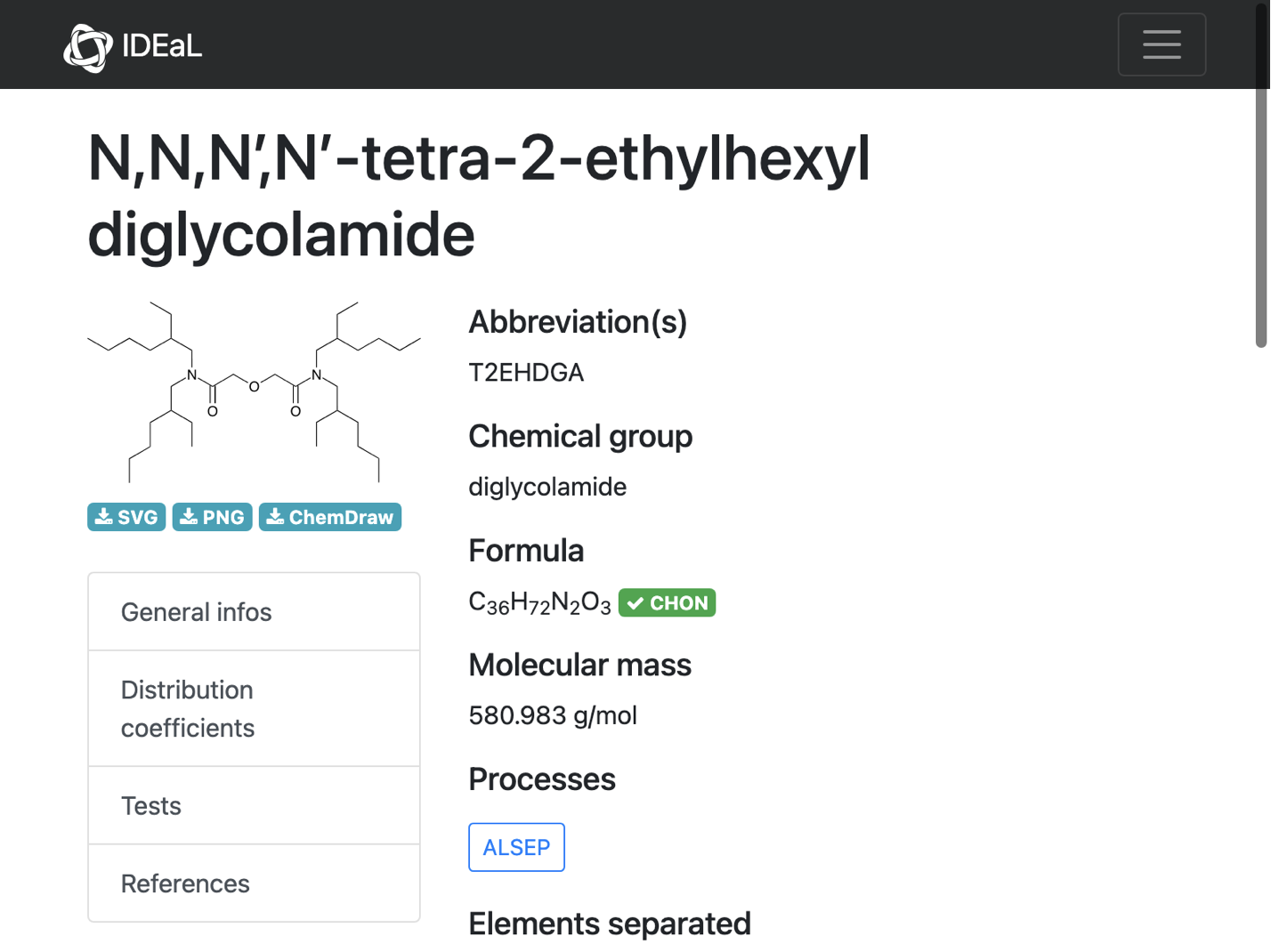
cdxMol = moleculeFromChemdraw[url]
formula = formulaExtract[url]
MoleculeValue[cdxMol, "MolecularFormulaString"] == formula
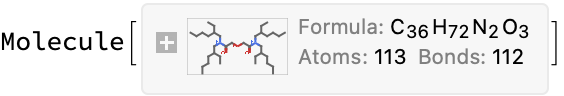
(*"C36H72N2O3"*)
(*True*)
solutions[[4]] = outputData[url, cdxMol]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/T2EHDGA", "abbreviation" -> "T2EHDGA", "SMILES" -> "CCCCC(CC)CN(CC(CC)CCCC)C(=O)COCC(=O)N(CC(CC)CCCC)CC(CC)CCCC", "InChI" -> "InChI=1S/C36H72N2O3/c1-9-17-21-31(13-5)25-37(26-32(14-6)22-18-10-2)35(39)29-41-30-36(40)38(27-33(15-7)23-19-11-3)28-34(16-8)24-20-12-4/h31-34H,9-30H2,1-8H3", "InChIKey" -> "MJJHOBMBUANRML-UHFFFAOYSA-N"|>*)
#5
url = problems[[5]]
URL[url]
WebImage[url]
(*"https://www.oecd-nea.org/ideal/extractants/ZS%20HDBP"*)

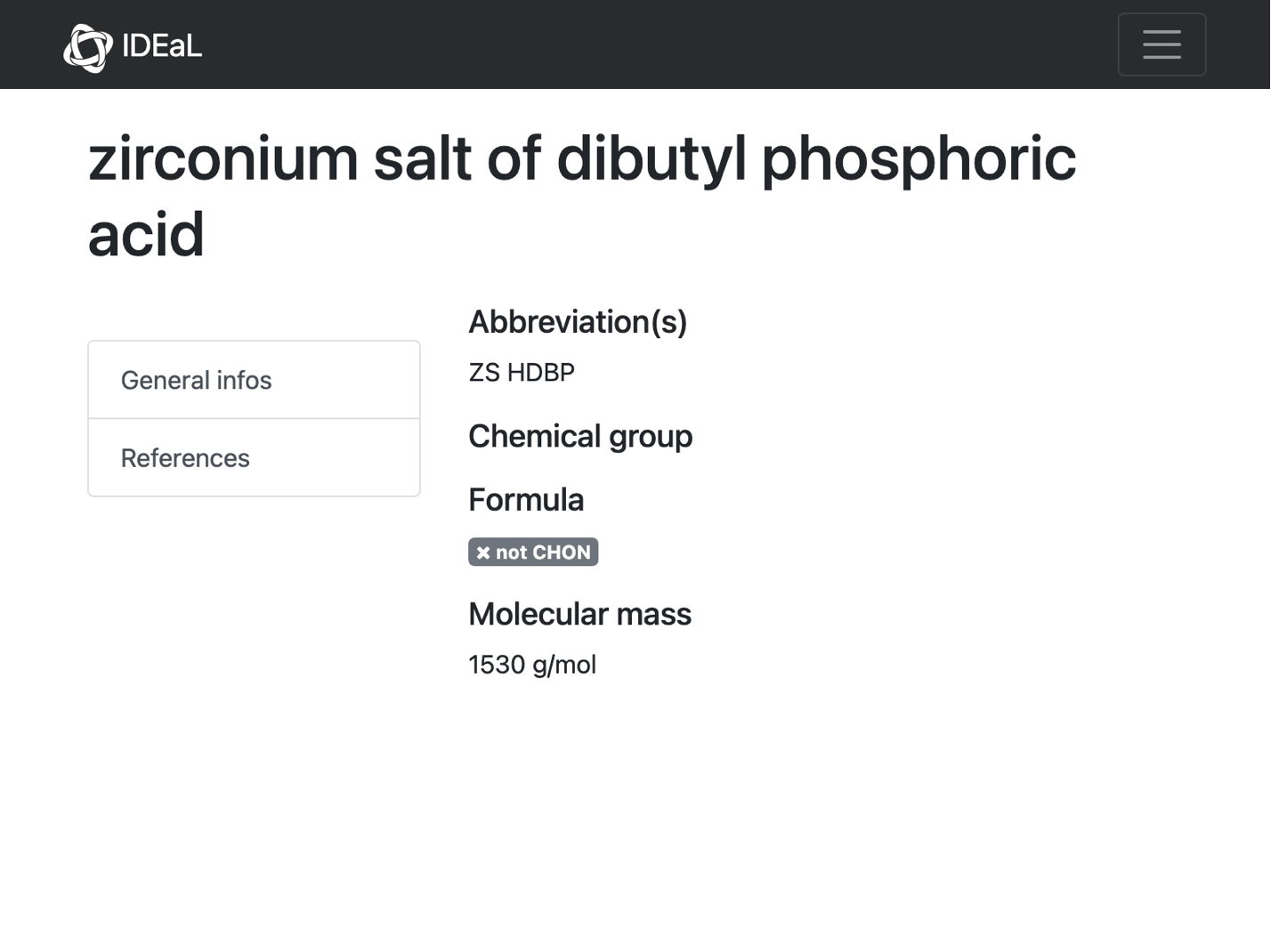
No D values and no chemdraw. So again, no reason to work too hard on this.
solutions[[5]] = outputData[url, Missing[]]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/ZS%20HDBP", "abbreviation" -> "ZS HDBP", "SMILES" -> Missing[], "InChI" -> Missing[], "InChIKey" -> Missing[]|>*)
Resolving the problem children
We have now resolved these error messages, obtaining the following resolutions:
solutions
(*{<|"url" -> "https://www.oecd-nea.org/ideal/extractants/9H", "abbreviation" -> "9H", "SMILES" -> "c1cc(C2=NN=C(c3ccncc3)NN2)ccn1", "InChI" -> "InChI=1S/C12H10N6/c1-5-13-6-2-9(1)11-15-17-12(18-16-11)10-3-7-14-8-4-10/h1-8H,(H,15,16)(H,17,18)", "InChIKey" -> "QWYDFEKBBFVMQW-UHFFFAOYSA-N"|>, <|"url" -> "https://www.oecd-nea.org/ideal/extractants/HONTA", "abbreviation" -> "HONTA", "SMILES" -> "CCCCCCCCN(CCCCCCCC)C(=O)CN(CC(=O)N(CCCCCCCC)CCCCCCCC)CC(=O)N(CCCCCCCC)CCCCCCCC", "InChI" -> "InChI=1S/C54H108N4O3/c1-7-13-19-25-31-37-43-56(44-38-32-26-20-14-8-2)52(59)49-55(50-53(60)57(45-39-33-27-21-15-9-3)46-40-34-28-22-16-10-4)51-54(61)58(47-41-35-29-23-17-11-5)48-42-36-30-24-18-12-6/h7-51H2,1-6H3", "InChIKey" -> "YTTXOOIRNCYALH-UHFFFAOYSA-N"|>, <|"url" -> "https://www.oecd-nea.org/ideal/extractants/SO3-Ph-BTBP", "abbreviation" -> "SO3-Ph-BTBP", "SMILES" -> Missing[], "InChI" -> Missing[], "InChIKey" -> Missing[]|>, <|"url" -> "https://www.oecd-nea.org/ideal/extractants/T2EHDGA", "abbreviation" -> "T2EHDGA", "SMILES" -> "CCCCC(CC)CN(CC(CC)CCCC)C(=O)COCC(=O)N(CC(CC)CCCC)CC(CC)CCCC", "InChI" -> "InChI=1S/C36H72N2O3/c1-9-17-21-31(13-5)25-37(26-32(14-6)22-18-10-2)35(39)29-41-30-36(40)38(27-33(15-7)23-19-11-3)28-34(16-8)24-20-12-4/h31-34H,9-30H2,1-8H3", "InChIKey" -> "MJJHOBMBUANRML-UHFFFAOYSA-N"|>, <|"url" -> "https://www.oecd-nea.org/ideal/extractants/ZS%20HDBP", "abbreviation" -> "ZS HDBP", "SMILES" -> Missing[], "InChI" -> Missing[], "InChIKey" -> Missing[]|>}*)
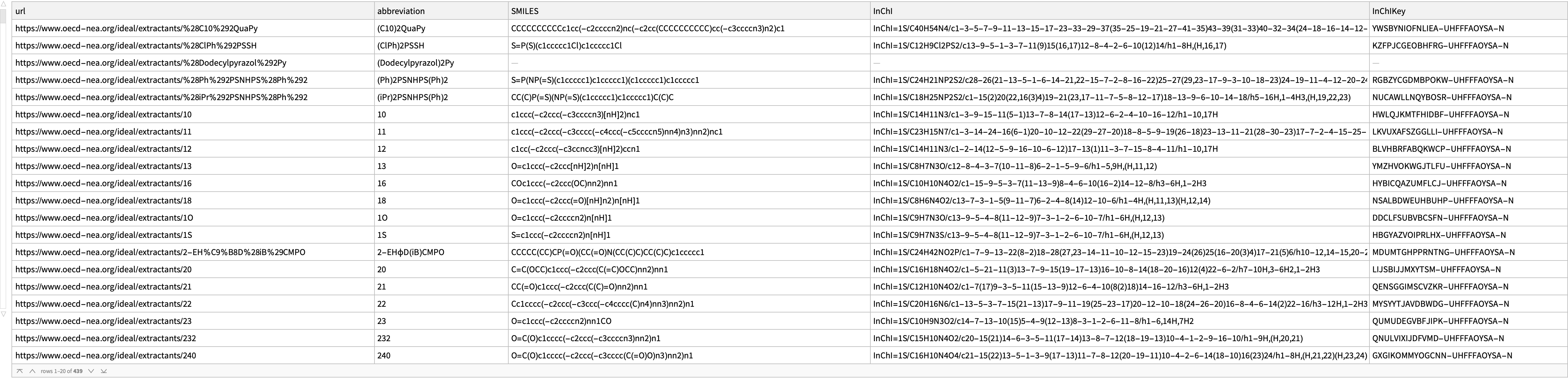
Keep the valid associations from the first case and append our resolved solutions`:
Dataset[
results2 = Join[ Select[AssociationQ]@results, solutions]]
Investigating the Missing entries
We still have many entries which did not give an overt failure, and yet we did not resolve a name. We must try to figure out what is going on here:
missingInfo = Select[MissingQ[#["SMILES"]] &]@results2;
Length[missingInfo]
(*105*)
An example
Take a look at thefirst example:
url = missingInfo[[1, "url"]]
WebImage[%]
info = compoundInformation[%%]
(*"https://www.oecd-nea.org/ideal/extractants/%28Dodecylpyrazol%292Py"*)
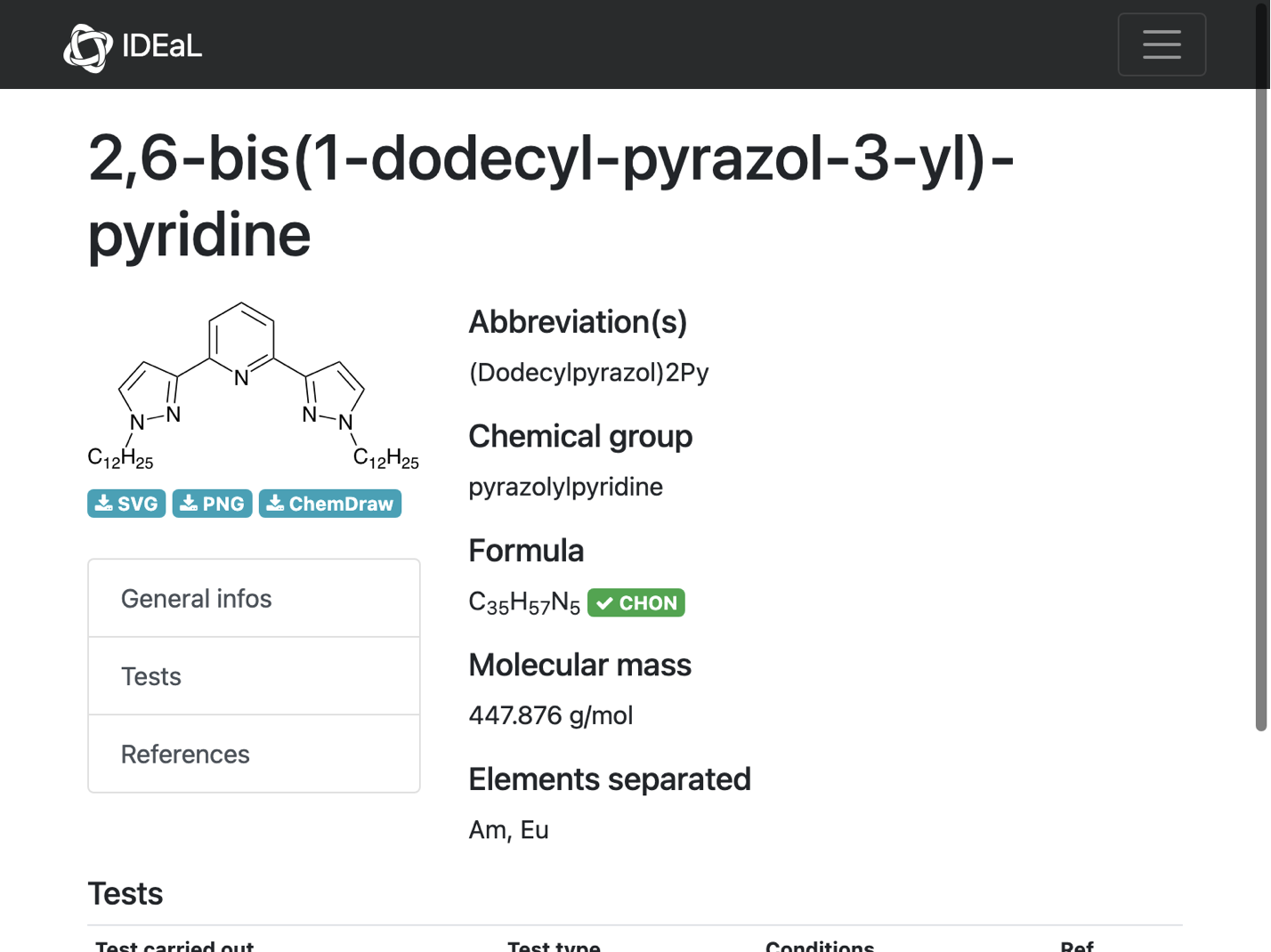
(*<|"compound_name" -> "2,6-bis(1-dodecyl-pyrazol-3-yl)-pyridine", "molecular_formula" -> "C35H57N5", "molecular_mass" -> "447.876"|>*)
This appears to be a case where the chemdraw is bad, but the name is fine; this tends to be a common error for many of these molecules which have large alkyl side chains (the ChemDraw structures are missing these entirely).
cdxMol = moleculeFromChemdraw[url]
iupacMol = interpretName[info]
moleculeConsistentQ[iupacMol, info]
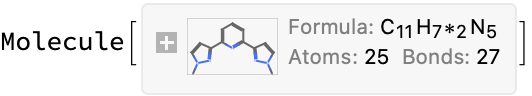
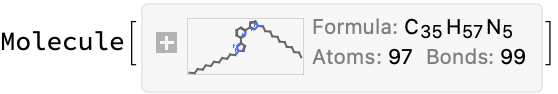
(*False*)
MoleculePlot[iupacMol]
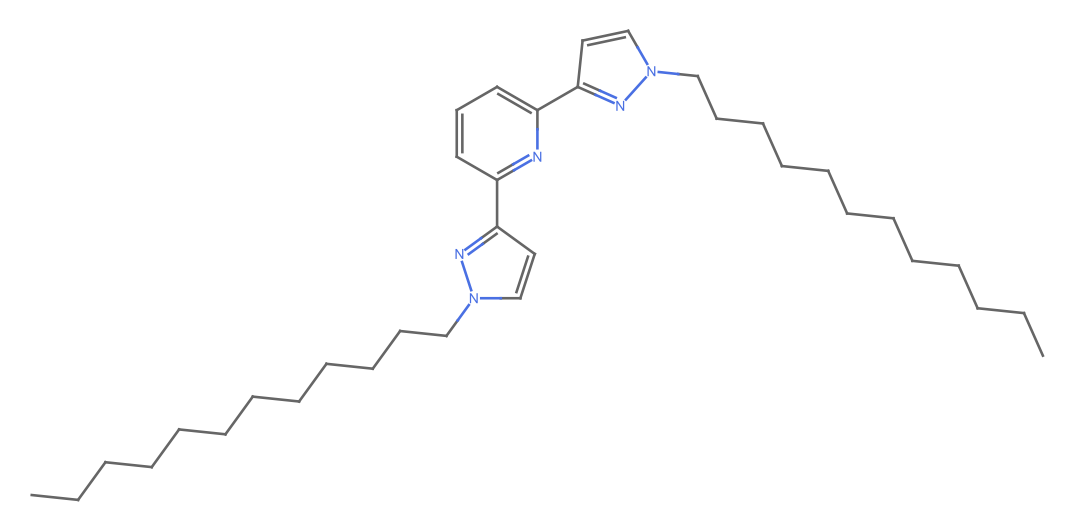
The structure appears to be correct and the chemical formula matches. But the posted molar mass on the website is wrong!
MoleculeValue[iupacMol, "MolarMass"]
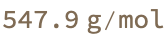
This suggests a more general problem elsewhere in the dataset which we shall need to investigate …
Where are the molecular weights inconsistent with the formulas?
massFormulaOnly[url_] := ImportString[#, "RawJSON"] & @ LLMFunction["What is the molecular formula and the molecular mass in the following text? Return only the formula and number with no units or other information. Return the results as a JSON dictionary with keys molecular_formula and molecular_mass.\n``",
ProgressReporting -> False]@
StringTake[#, UpTo[1500]] &@Import[url, "Plaintext"]
computeMassFromFormula[info_Association] := ChemicalFormula[info["molecular_formula"]]["MolecularMass"] // QuantityMagnitude
consistentMassQ[info_Association] := With[
{statedMass = Interpreter[ "Number"]@info["molecular_mass"],
computedMass = computeMassFromFormula[info] },
Round[statedMass] == Round[computedMass]]
inconsistentMass[url_] := With[
{info = massFormulaOnly[url]},
If[! consistentMassQ[info],
<|"url" -> url,
"formula" -> info["molecular_formula"],
"stated_mass" -> Interpreter["Number"][info["molecular_mass"]],
"compute_mass" -> computeMassFromFormula[info]
|>,
Nothing
]
]
(*demo*)
inconsistentMass[url]
(*<|"url" -> "https://www.oecd-nea.org/ideal/extractants/%28Dodecylpyrazol%292Py", "formula" -> "C35H57N5", "stated_mass" -> 447.876, "compute_mass" -> 547.9|>*)
As usual, we will parallelize on the HTTP requests / API calls to speed things up a bit (most of the time is spent having our computer wait for responses). Some errors arise for gateway failures or other problems:
inconsistencies = ParallelMap[inconsistentMass, Lookup[missingInfo, "url"]];
Dataset[%]

We will just drop the gateway errors or cases of missing formulas. I have contacted a representative from OECD-NEA.org (12 Oct 2023) to make them aware of these discrepancies. Alas, it appears that this accounts for only six of our problems
Dataset@Select[AssociationQ]@inconsistencies
Export["2023.10.12_inconsistent_formula_masses_ideal.xlsx", %]
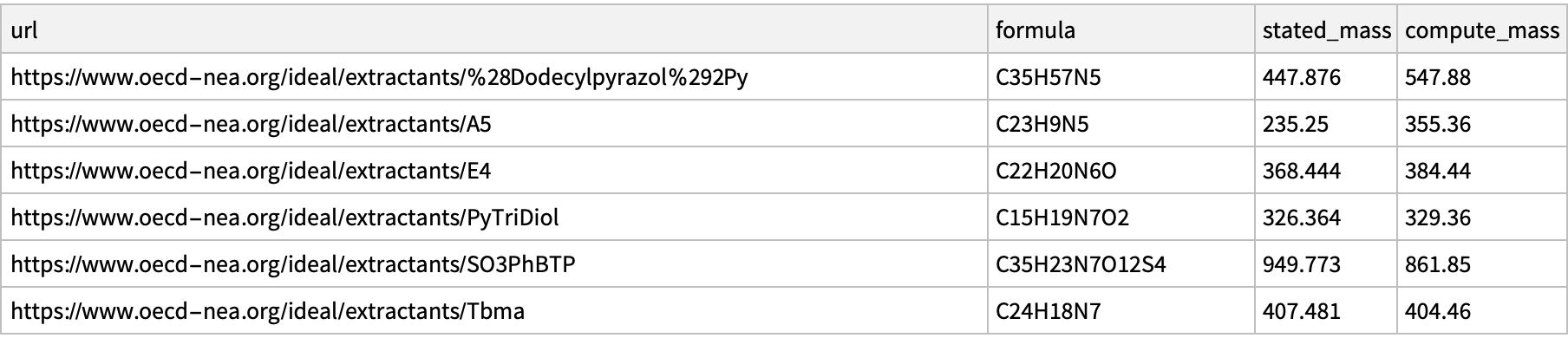
(*"2023.10.12_inconsistent_formula_masses_ideal.xlsx"*)
Retrieve the structures for these inconsistent results:
foundMols = moleculeFromChemdraw /@ Lookup[ Select[AssociationQ]@inconsistencies, "url"]
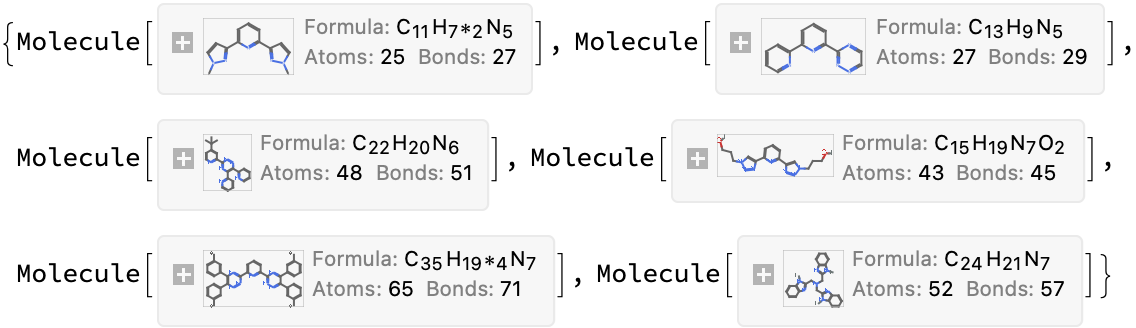
How does this compare to the listed formula?
Lookup["formula"]@Select[AssociationQ]@inconsistencies
(*{"C35H57N5", "C23H9N5", "C22H20N6O", "C15H19N7O2", "C35H23N7O12S4", "C24H18N7"}*)
Comment: Only #4 has a chemdraw file consistent with the molecular formula, but given all the other discrepancies I think this should be investigated further.
Investigating other problems
Other cases appear to have totally inconsistent information. Here is one example:
WebImage@missingInfo[[3, "url"]]
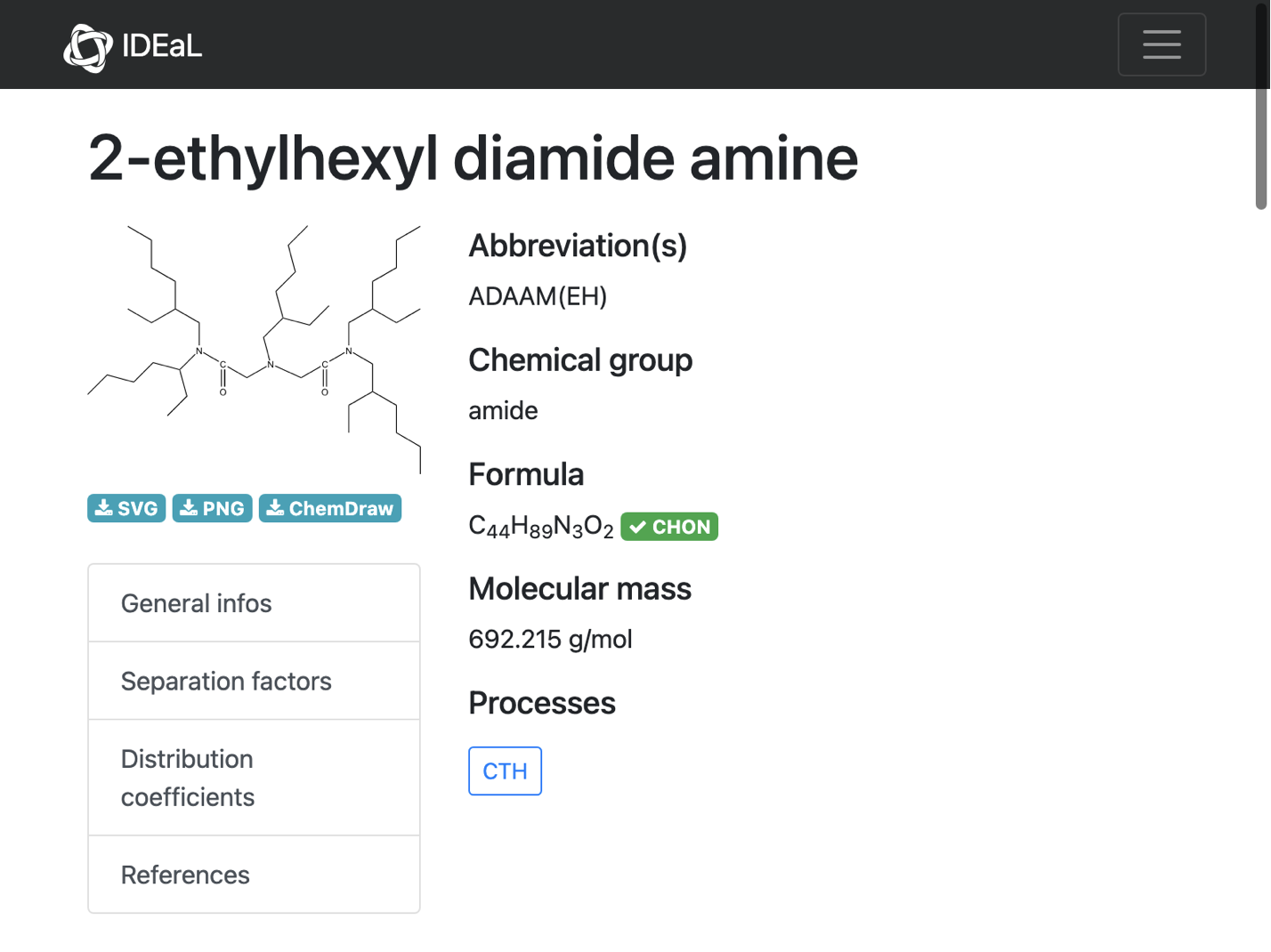
The CDX file is suspicious (missing atoms?), and the stated name is unresolvable:
cdxMol = moleculeFromChemdraw@missingInfo[[3, "url"]]
info = compoundInformation@missingInfo[[3, "url"]]
iupacMol = Molecule@info["compound_name"]

(*<|"compound_name" -> "2-ethylhexyl diamide amine", "molecular_formula" -> "C44H89N3O2", "molecular_mass" -> "692.215"|>*)

(*Molecule["2-ethylhexyl diamide amine"]*)
You can also try to OCR the image, but this is also inconsistent with the stated molecular formula:
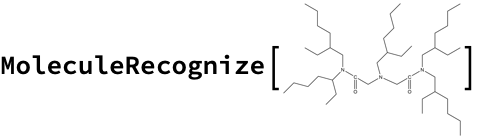
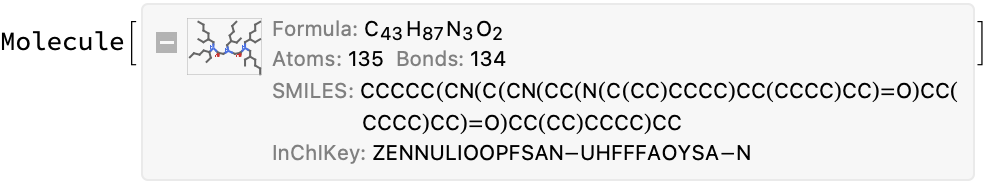
Conclusion: There is no simple solution here. Lots of inconsistent results. This is one where a human will have to go back to the original reference papers to sort out.
How many of these problem entries do we actually care about?
We do not care about missing entries unless they contain distribution coefficient data. How many of these have D values?
containsDValuesQ[url_] :=
StringContainsQ["Distribution coefficients", IgnoreCase -> True]@Import[url, "Plaintext"]
(*demo*)
containsDValuesQ@missingInfo[[1, "url"]]
(*False*)
Now apply to the entire list:
missingInfo2 = Append[#, "containsDValuesQ" -> containsDValuesQ[#["url"]]] & /@ missingInfo;
Counts@Lookup[missingInfo2, "containsDValuesQ"]
(*<|False -> 30, True -> 75|>*)
Conclusion: Too bad. Looks like we have 75 cases that will need attention. Write them out to a spreadsheet and dispatch someone to get to work…
Export["2023.10.12_inconsistent_entries_with_D_values.xlsx",
Select[#["containsDValuesQ"] &]@Dataset@missingInfo2];
Conclusion
Even so, we still have a large number of the results that can be used:
Export["2023.10.12_completed_ideal_molecules.xlsx",
Select[! MissingQ[#["SMILES"]] &]@Dataset@results2];
Export["2023.10.12_missing_entries.xlsx", Dataset[missingInfo2]];
You can download the final spreadsheet file of correct values here. Merge this with your scraped data tables (indexing on the the URL or abbreviation fields) and you are off to the races.
You can also download the spreadsheet of unresolved entries with D values and the spreadsheet containing the subset of these with inconsistent molecular formulas and masses. There is also a complete list of all unresolved entries
The Sequel: The Revenge of ChemDraw
Upon further poking on this, it turns out that we can hack around this problem by AppleScripting Chemdraw to export SMILES strings. We treat this in depth in the next post…
ToJekyll["Parsing molecular identifiers from the IDEaL Database", "llm mathematica science"]
-
Looking back at my usage log, I spent about $0.40 USD in total for all of the calls made in writing, debugging, and using this blog post. ↩