Accessing the Ginkgo Protein Foundation Model API
[science proteins biology llm ml machinelearning mathematica In a previous post we discussed various protein embedding models. Although Mathematica 14.2 added some limited support for ESMAtlas, the current version only supports retrieving embedding vectors for entries with a MGnifyID, not arbitrary user-specified sequences. An alternative is to use the new Ginkgo Bioworks API which can perform both masked generation and embedding calculations using the ESM2 model as well as their own proprietary Ginkgo-AA0 model. A short tour of how to access the Ginkgo API to compute protein embeddings…
We will just make up a peptide sequence here (stored as a string, rather than anything fancy like BioSequence):
example = "MKLAKRVSALTPSTTLAITAKAAPD"
(* "MKLAKRVSALTPSTTLAITAKAAPD" *)
We will also make an example for mask filling demonstration. In general, it uses the ESMTokenizer, which accepts uppercase characters representing amino acids as well as the special tokens <unk>, <pad>, <cls>, <mask>, and <eos>. We will replace one amino acid with a mask:
maskExample = StringReplacePart["<mask>", {5, 5}]@ example (* mask amino acid 5 *)
(As an aside, most of the code examples they provide are centered around using the mask filling capability, but we will be focusing on the protein embedding capability instead. They also provide a Python client, which is just a wrapper around some simple HTTP calls, as we shall see below.)
Create an account at GinkgoBioworks.ai. After you log in, you will see a page that says your API key. We will store the API key in the secure persistent datastore for later use:
SystemCredential["GinkgoAPIKey"] = (* YOUR SECRET KEY *)
On HTTP Requests in Mathematica
HTTPRequest creates symbolic representations of an HTTP request (but does not send it).
URLRead will evaluate the request and return a symbolic HTTPResponse which contains status code and other information. This is probably more informative, particularly for sophisticated error handling. There is also some capability to provide an element or list of elements to retrieve from the response body.
URLExecute will evaluate the HTTPRequest and return the response body interpreted as a Wolfram language expression (typically as a list of rules).
Single embedding calculation:
We will begin by defining a function which sends the request to the API:
ginkgoModelRequest[sequence_String,
requestType_String : "EMBEDDING", (* or FILL_MASK *)
model_String : "ginkgo-aa0-650M"] := HTTPRequest[
URL[ "https://api.ginkgobioworks.ai/v1/transforms/run"],
<|
Method -> "POST",
"ContentType" -> "application/json",
"Headers" -> { "x-api-key" -> SystemCredential["GinkgoAPIKey"]},
"Body" -> ExportString[#, "JSON"] &@<|
"model" -> model,
"text" -> sequence,
"transforms" -> { <|"type" -> requestType|>}|>
|>]
Then execute the request; it should return a jobId and a result URL to check for the result when it is completed:
apiOutput = URLExecute@ ginkgoModelRequest[ex]
(*
{"jobId" -> "40efe9e2-3fa9-4fa4-9ef7-0a7ec3a3f0c1",
"result" -> "https://api.ginkgobioworks.ai/v1/transforms/run?jobId=40efe9e2-3fa9-4fa4-9ef7-0a7ec3a3f0c1",
"status" -> "PENDING"}
*)
Query the result URL to obtain the result using a simple GET request:
gingkoResultRequest[url_String | url_URL] := HTTPRequest[
url,
<|
Method -> "GET",
"ContentType" -> "application/json",
"Headers" -> { "x-api-key" -> SystemCredential["GinkgoAPIKey"]}
|>]
Check the output?
result = URLExecute@ gingkoResultRequest@ Lookup["result"]@ apiOutput;
Short[%, 7]
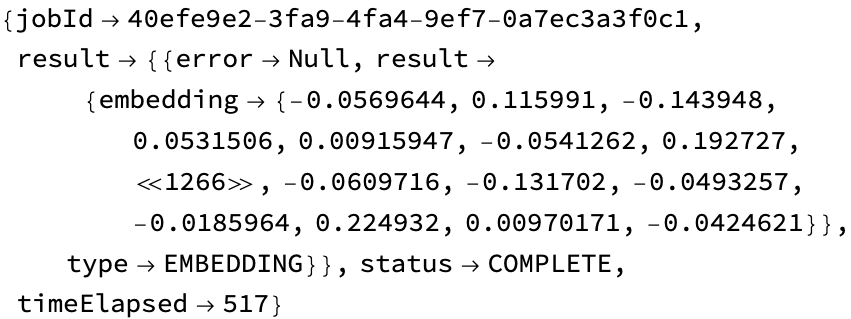
You should probably confirm that the status is COMPLETE before doing anything (in practice, this calculation runs very quickly):
Lookup["status"]@ result
(*"COMPLETE"*)
The embedding is stored in a dictionary-list-dictionary-dictionary structure, so it is convenient to pull it out:
extractEmbeddingResult = Query["result", 1, "result", "embedding"];
Short@ extractEmbeddingResult@ result (* pull out result *)
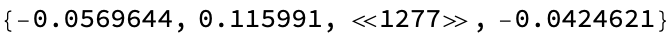
It is straightforward to generalize this to mask filling as well:
extractMaskResult = Query["result", 1, "result", "sequence"];
URLExecute@ ginkgoModelRequest[maskExample, "FILL_MASK"]
Pause[2] (* wait two seconds *)
URLExecute@ gingkoResultRequest@Lookup["result"]@ %%
extractMaskResult@%
(*
{"jobId" -> "b40724d4-8875-478b-86b7-d4ac345514d0",
"result" -> "https://api.ginkgobioworks.ai/v1/transforms/run?jobId=b40724d4-8875-478b-86b7-d4ac345514d0",
"status" -> "PENDING"}
*)
(*
{"jobId" -> "b40724d4-8875-478b-86b7-d4ac345514d0",
"result" -> { {"error" -> Null, "result" -> {"sequence" -> "MKLAARVSALTPSTTLAITAKAAPD"}, "type" -> "FILL_MASK"}},
"status" -> "COMPLETE", "timeElapsed" -> 684}
*)
(* "MKLAARVSALTPSTTLAITAKAAPD" *)
Put it all together, polling the server until it is complete. (This is not the most robust code, but maybe it is enough for a typical scientific user)
gingkoLookup[sequence_String,
requestType_String : "EMBEDDING", (* or "FILL_MASK" *)
model_String : "ginkgo-aa0-650M"] := Module[
{resultRequest, result, iterations = 0},
(* generate the GET response needed to retrieve the result *)
resultRequest = gingkoResultRequest@Lookup["result"]@
URLExecute@ ginkgoModelRequest[sequence, requestType, model];
(* wait a second between queries, and wait a maximum of 30 seconds before failing *)
Until[
(StringMatchQ["COMPLETE"]@Lookup["status"]@result) || (iterations > 30),
Pause[1];
result = URLExecute[resultRequest];
iterations++;
];
(* extract the relevant result *)
Switch[requestType,
"EMBEDDING", extractEmbeddingResult@ result,
"FILL_MASK", extractMaskResult@ result
]]
Batch calculations
One typically wants to perform the evaluation for a batch of sequences at once. Poking the API for each one individually would be annoying, so instead we will use the batch capability. Begin by defining a trivial example:
examples = {example, example};
Overload the request function to catch a list argument containing multiple sequences. Inside the function, generate a list of inputs and the endpoint URL is a bit different, but the changes are relatively minor. (The documentation does not indicate what the maximum batch size should be, but an example shows batches of 10, so maybe we can consider that as a guide; in subsequent testing I had no problem with batches of 100 queries, although they can take 1-2 minutes to complete, depending on the model.)
ginkgoModelRequest[sequences_List, (*overload to take a list *)
requestType_String : "EMBEDDING", (* or FILL_MASK *)
model_String : "ginkgo-aa0-650M"] :=
With[
{requestBody =
<|"model" -> model, "text" -> #, "transforms" -> {<|"type" -> requestType|>} |> & /@ sequences},
HTTPRequest[
URL[ "https://api.ginkgobioworks.ai/v1/batches/transforms/run"],
<|
Method -> "POST",
"ContentType" -> "application/json",
"Headers" -> { "x-api-key" -> SystemCredential["GinkgoAPIKey"]},
"Body" -> ExportString[<|"requests" -> requestBody|>, "JSON"]
|>]]
When we execute this eq, we again get back a result:
response = URLExecute@ginkgoModelRequest[examples]
(*
{"batchId" -> "32ec5701-5b89-415f-b818-192960938a18",
"jobIds" -> {"09f00bb0-3a8b-4c6d-902c-57a1f25d4a72", "8b45326e-269e-4e1f-b7a2-cb86c49c69b6"},
"result" -> "https://api.ginkgobioworks.ai/v1/batches/transforms/run?batchId=32ec5701-5b89-415f-b818-192960938a18",
"status" -> "PENDING"}
*)
The trick is that we have to keep track of both the jobIds and the result URL; there are some notes on the python API readme about how the results can get returned in various orders, so we will maintain a list of both of these:
jobIDs = Lookup["jobIds"]@response
url = Lookup["result"]@response
(* {"09f00bb0-3a8b-4c6d-902c-57a1f25d4a72", "8b45326e-269e-4e1f-b7a2-cb86c49c69b6"} *)
(* "https://api.ginkgobioworks.ai/v1/batches/transforms/run?batchId=32ec5701-5b89-415f-b818-192960938a18" *)
Go ahead and pull the results (presumably complete):
(results = URLExecute@gingkoResultRequest[url]) // Short[#, 8] &
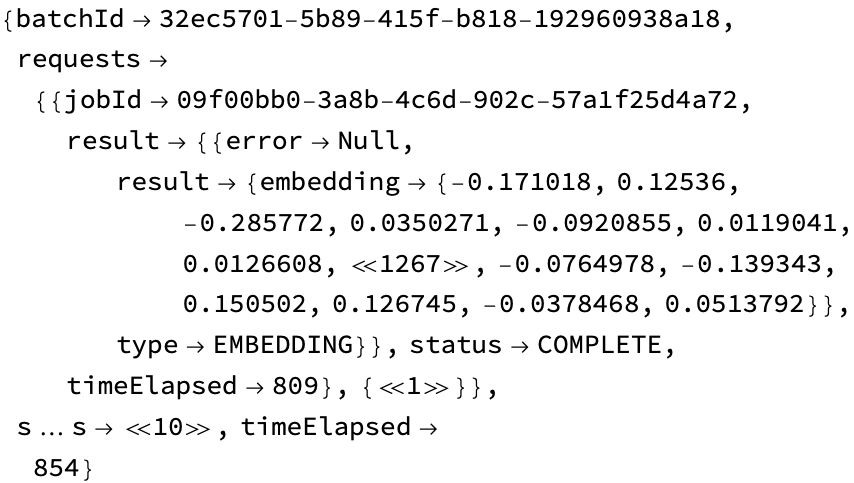
Extract the ids:
ids = Query["requests", All, "jobId"]@ results
(* {"09f00bb0-3a8b-4c6d-902c-57a1f25d4a72", "8b45326e-269e-4e1f-b7a2-cb86c49c69b6"} *)
And the corresponding embeddings:
(embeddings = Query["requests", All, "result", 1, "result", "embedding", All]@ results) // Short[#, 8] &

We can batch process the response to match the sequences to the resulting embeddings ; this is also an excuse to get acquainted with the new Tabular functionality in Mathematica 14.2:
match[sequences_, response_, results_] := With[
{jobs = Lookup[response, "jobIds"],
id2Embedding = AssociationThread[
Query["requests", All, "jobId"]@results ->
Query["requests", All, "result", 1, "result", "embedding", All]@ results]},
ToTabular[
{TabularColumn[sequences],
TabularColumn[id2Embedding /@ jobs]},
"Columns",
<|"ColumnKeys" -> {"sequence", "embedding"}|>]]
match[examples, response, results] // Short[#, 8] &
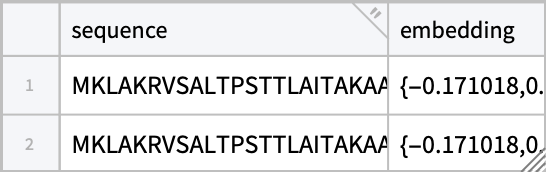
(Generalization to handle mask filling is left as an exercise for the reader.)
So you’ve got a bunch of embeddings to store
Make it easy to share, and do what the cool kids are doing: Parquet… added in Mathematica 14.2
ToJekyll["Accessing the Ginkgo Protein Foundation Model API",
"science proteins biology llm ml machinelearning"]