Semantic Article Retrieval from CrossRef with LLM Embeddings
[science llm machinelearning ml mathematica Suppose you wanted to gather all of the scholarly articles related to a topic (e.g., constructing a database of f-element solvent separations)? You could do a keyword search, but that might include many useless papers or fail to include papers with the exact specified keywords in the title or abstract. A modern approach would be to try to find articles that are semantically close by generating an embedding vector that represented the document (using your favorite foundational language model) and then perform some sort of clustering or classification to retrieve documents of interest–this might be as simple as just finding documents where the vectors are close enough (below some threshold). A code sketch demonstration of how to conduct this process (and some of the complications that may arise)…
Retrieving articles from CrossRef
CrossRef provides free metadata (title, abstract, doi, reference, authors, etc.) on scholarly articles. Mathematica has a built-in functionality for CrossRef API queries (or roll your own REST-API calls), which will return batches of 35 articles at a time, based upon a word search.
examples = ServiceExecute["CrossRef", "WorksDataset",
{"Query" -> "actinide separation", "TypeID" -> "journal-article"}];
examples[[1]]
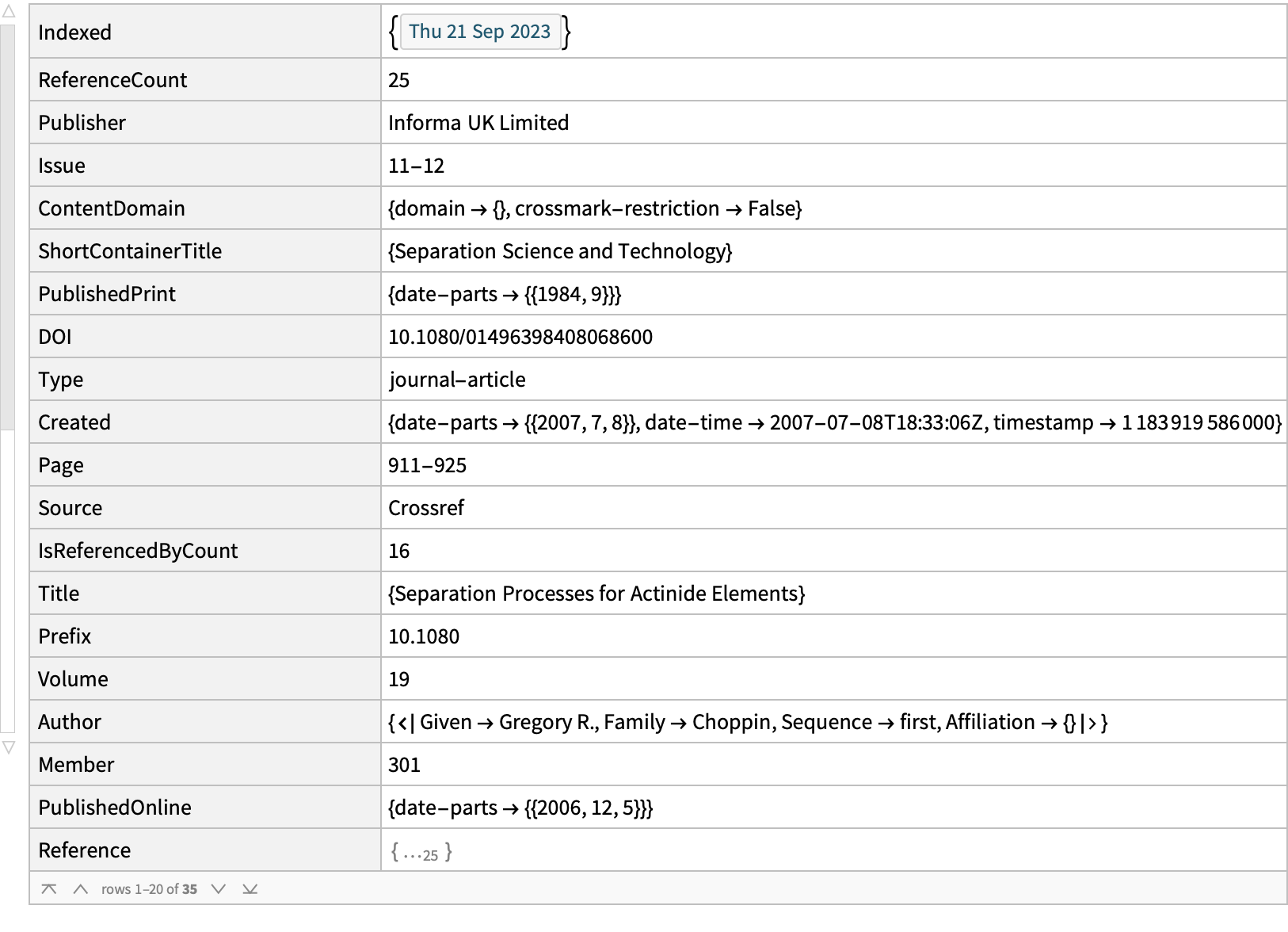
We would have to do pages of retrievals, define keywords, etc., which is exactly what we do not want to do. Instead we want all titles and abstracts, and will devise our own way of grouping them. For big comprehensive projects involving bulk queries, CrossRef prefers that you download the complete torrent of their data and process that locally, supplementing it with API queries for newer information (one can provide date restrictions in the API). Transmission is a pretty decent bit-torrent client. The April 2023 CrossRef source is: [https://academictorrents.com/details/d9e554f4f0c3047d9f49e448a7004f7aa1701b69]
The torrent is comprised of individual gziped JSON files (of 5000 articles each). Because the files are separate, you do not have to download it all to do a demonstration; I paused the download after the first few gigabytes. Begin by reading in one of these files as an example:
cr = Import["~/Downloads/crossref/April 2023 Public Data File from Crossref/20299.json.gz", "RawJSON"]["items"]
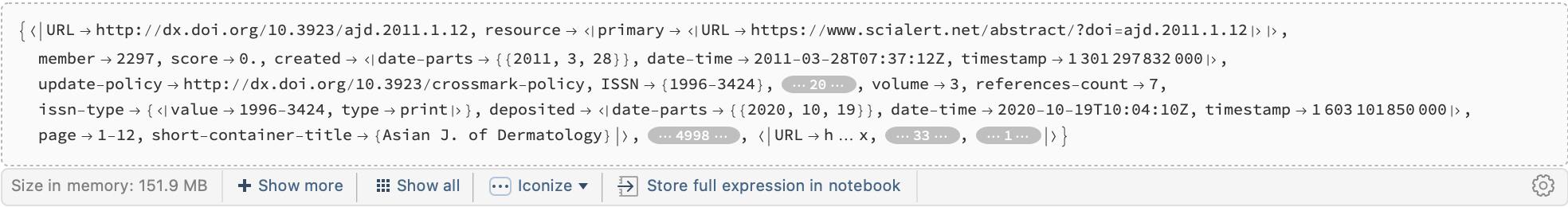
Here is an example of what might be in a record:
cr[[100]]
(*<|"URL" -> "http://dx.doi.org/10.1017/s0022046900024921", "resource" -> <|"primary" -> <|"URL" -> "https://www.cambridge.org/core/product/identifier/S0022046900024921/type/journal_article"|>|>, "member" -> "56", "score" -> 0., "created" -> <|"date-parts" -> 2011, "date-time" -> "2011-03-28T04:23:29Z", "timestamp" -> 1301286209000|>, "license" -> {<|"start" -> <|"date-parts" -> 2011, "date-time" -> "2011-03-25T00:00:00Z", "timestamp" -> 1301011200000|>, "content-version" -> "unspecified", "delay-in-days" -> 10675, "URL" -> "https://www.cambridge.org/core/terms"|>}, "ISSN" -> {"0022-0469", "1469-7637"}, "container-title" -> {"The Journal of Ecclesiastical History"}, "issued" -> <|"date-parts" -> 1982|>, "issue" -> "1", "prefix" -> "10.1017", "reference-count" -> 0, "indexed" -> <|"date-parts" -> 2022, "date-time" -> "2022-04-01T19:22:16Z", "timestamp" -> 1648840936407|>, "author" -> {<|"given" -> "Owen", "family" -> "Chadwick", "sequence" -> "first", "affiliation" -> {}|>}, "DOI" -> "10.1017/s0022046900024921", "is-referenced-by-count" -> 0, "published" -> <|"date-parts" -> 1982|>, "published-print" -> <|"date-parts" -> 1982|>, "alternative-id" -> {"S0022046900024921"}, "subject" -> {"Religious studies", "History"}, "published-online" -> <|"date-parts" -> 2011|>, "content-domain" -> <|"domain" -> {}, "crossmark-restriction" -> False|>, "title" -> {"Theologische Realenzyklopa"die. Edited by G. Krause and G. Mu"ller. v, 5, pp. 164; vi, 1\[Dash]5, pp. 785 + plates and drawings. Berlin\[LongDash]New York: Walter de Gruyter, 1980. Subscription price per fascicle DM 38."}, "link" -> {<|"URL" -> "https://www.cambridge.org/core/services/aop-cambridge-core/content/view/S0022046900024921", "content-type" -> "unspecified", "content-version" -> "vor", "intended-application" -> "similarity-checking"|>}, "source" -> "Crossref", "type" -> "journal-article", "publisher" -> "Cambridge University Press (CUP)", "journal-issue" -> <|"issue" -> "1", "published-print" -> <|"date-parts" -> 1982|>|>, "volume" -> "33", "references-count" -> 0, "issn-type" -> {<|"value" -> "0022-0469", "type" -> "print"|>, <|"value" -> "1469-7637", "type" -> "electronic"|>}, "deposited" -> <|"date-parts" -> 2019, "date-time" -> "2019-05-25T15:11:50Z", "timestamp" -> 1558797110000|>, "language" -> "en", "page" -> "167-168", "short-container-title" -> {"J. Eccles. Hist."}|>*)
Comment: Not all items have abstracts! (the above is an example) Including an abstract is optional and up to the publishers, so the solution we build should not rely upon having this type of data.
You might be tempted to do an initial screening by subject type:
cr[[6, "subject"]]
(*{"General Engineering"}*)
However, this was only added on an experimental basis and is not officially supported. In practice, many of these do not have a subject, and most of them don’t have an abstract either…
Count["MISSING"]@ Lookup[cr, "subject", "MISSING"]
Count["MISSING"]@ Lookup[cr, "abstract", "MISSING"]
(*1132*)
(*4239*)
So we will confine our efforts to the title and abstract (if present). Here’s an example of one that seems complete:
cr[[6, {"title", "abstract", "DOI"}]]
(*<|"title" -> {"Nanolayered CrN/TiAIN Coatings Synthesized by Ion Plating Technology for Tribological Application on Piston Rings"}, "abstract" -> "<jats:p>By using the ion plating technology,the multilayered CrN/TiAlN as well as monolayered CrN and TiAlN coatings were made on the suface of piston rings to improve its tribological properties and increase service life using Cr and Ti50Al50 alloy cathodes. EDS analysis showed that the main compositions of CrN/TiAlN coatings are Cr30.61%,Ti 20.42%,Al 13.88% and N 35.10%. The preferred orientation was changed from (111) in CrN and TiAlN monolayered coatings to (220) plane in the multilayered CrN/TiAlN coatings. The multilayered CrN/TiAlN coatings had smaller crystallite size than the monolayered CrN coatings. The multilayered TiAlN/CrN coating with rotational speed at 1.5 rpm exhibited the highest H3/E2 ratio value of 0.23 GPa, indicating the best resistance to plastic deformation, among the studied CrN, TiAlN and multilayered TiAlN/ CrN coatings. The CrAlTiN composite films performs better than binary CrN as well as the Cr plating in teams of hardness and wear resistance at high temperature.</jats:p>", "DOI" -> "10.4028/www.scientific.net/amr.216.430"|>*)
Additionally, not all articles (especially non-English articles) have a title. Such records have original-title (in the source language) and then container-title and short-container-title provided. Here is an example:
cr[[474]]
(*<|"URL" -> "http://dx.doi.org/10.3902/jnns.12.135", "resource" -> <|"primary" -> <|"URL" -> "http://www.jstage.jst.go.jp/article/jnns/12/2/12_2_135/_article/-char/ja/"|>|>, "member" -> "2266", "score" -> 0., "created" -> <|"date-parts" -> 2011, "date-time" -> "2011-03-28T06:07:15Z", "timestamp" -> 1301292435000|>, "ISSN" -> {"1883-0455", "1340-766X"}, "container-title" -> {"The Brain & Neural Networks"}, "issued" -> <|"date-parts" -> 2005|>, "issue" -> "2", "prefix" -> "10.3902", "reference-count" -> 0, "indexed" -> <|"date-parts" -> 2022, "date-time" -> "2022-03-29T07:03:20Z", "timestamp" -> 1648537400751|>, "DOI" -> "10.3902/jnns.12.135", "is-referenced-by-count" -> 0, "original-title" -> {"\:6c96\:7e04\:8a08\:7b97\:795e\:7d4c\:79d1\:5b66\:30b3\:30fc\:30b9\:306b\:53c2\:52a0\:3057\:3066"}, "published" -> <|"date-parts" -> 2005|>, "published-print" -> <|"date-parts" -> 2005|>, "content-domain" -> <|"domain" -> {}, "crossmark-restriction" -> False|>, "title" -> {""}, "source" -> "Crossref", "type" -> "journal-article", "publisher" -> "Japanese Neural Network Society", "journal-issue" -> <|"issue" -> "2", "published-print" -> <|"date-parts" -> 2005|>|>, "volume" -> "12", "references-count" -> 0, "issn-type" -> {<|"value" -> "1883-0455", "type" -> "electronic"|>, <|"value" -> "1340-766X", "type" -> "print"|>}, "deposited" -> <|"date-parts" -> 2021, "date-time" -> "2021-05-20T18:31:26Z", "timestamp" -> 1621535486000|>, "language" -> "ja", "page" -> "135-136", "short-container-title" -> {"The Brain & Neural Networks", "Nihon Shinkei Kairo Gakkaishi"}|>*)
Comment: Also notice how this record lists the key as DOI instead of doi on other references. So we need both…
If we put this into production then we may want to handle this in a smarter way (e.g., grabbing all of the alternates and only keeping the first one that appears). If there is no abstract, then we default to any title that is present.
ClearAll[filter, articles]
(*lookup multiple properties in priority order, so that at least one works*)
redundantLookup[record_Association, keys_List, missing_ : "Missing"] :=First@DeleteCases[""]@Flatten@Append["Missing"]@Lookup[record, keys, Nothing]
filter[record_Association] := Association[
"title" -> redundantLookup[record, {"title", "container-title", "original-title", "short-container-title"}], "abstract" -> redundantLookup[record, {"abstract", "title"}],
"doi" -> redundantLookup[record, {"doi", "DOI"}]
]
articles = filter /@ cr;
This gives us some nicely structured data (a few examples):
articles[[{1, 6, 474}]]
(*{<|"title" -> "Evaluation of Thyroid Hormone Abnormalities and Thyroid Autoantibodies in Chronic Idiopathic Urticaria and Alopecia Areata Egyptian Patients", "abstract" -> "Evaluation of Thyroid Hormone Abnormalities and Thyroid Autoantibodies in Chronic Idiopathic Urticaria and Alopecia Areata Egyptian Patients", "doi" -> "10.3923/ajd.2011.1.12"|>, <|"title" -> "Nanolayered CrN/TiAIN Coatings Synthesized by Ion Plating Technology for Tribological Application on Piston Rings", "abstract" -> "<jats:p>By using the ion plating technology,the multilayered CrN/TiAlN as well as monolayered CrN and TiAlN coatings were made on the suface of piston rings to improve its tribological properties and increase service life using Cr and Ti50Al50 alloy cathodes. EDS analysis showed that the main compositions of CrN/TiAlN coatings are Cr30.61%,Ti 20.42%,Al 13.88% and N 35.10%. The preferred orientation was changed from (111) in CrN and TiAlN monolayered coatings to (220) plane in the multilayered CrN/TiAlN coatings. The multilayered CrN/TiAlN coatings had smaller crystallite size than the monolayered CrN coatings. The multilayered TiAlN/CrN coating with rotational speed at 1.5 rpm exhibited the highest H3/E2 ratio value of 0.23 GPa, indicating the best resistance to plastic deformation, among the studied CrN, TiAlN and multilayered TiAlN/ CrN coatings. The CrAlTiN composite films performs better than binary CrN as well as the Cr plating in teams of hardness and wear resistance at high temperature.</jats:p>", "doi" -> "10.4028/www.scientific.net/amr.216.430"|>, <|"title" -> "The Brain & Neural Networks", "abstract" -> "Missing", "doi" -> "10.3902/jnns.12.135"|>}*)
Generate title embeddings
We are now going to take the text and generate embeddings–i.e., convert the text into a vector of numbers that we can then compare to identify similar articles. (If this is a new idea to you, then the OpenAI documentation is a nice starting place on how to use text embeddings)
If you are lazy, have an internet connection, and do not mind giving OpenAI your money, then you could just call their API and be done with it. There are some practical limits in terms of how many tokens you can provide in each batch: to be conservative I break this up into 4 batches of 1250 titles each (submitting each batch to the API in parallel); this costs us about $0.03 USD in total. As an aside, the OpenAI Terms of Use permits you to share your embeddings freely (user owns model outputs), so maybe somebody has already done this for us…(n.b. as of 09 Oct 2023, I am not aware of this existing):
responses = ParallelMap[
ServiceExecute["OpenAI", "Embedding", {"Input" -> #}] &,
Partition[ Lookup[articles, "title"], UpTo[1250]]];
openAITitleEmbeddings = Join @@ Table[ ( responses[[i, "data"]][[All, "embedding"]] ), {i, 4}];
Dimensions[%]
(*{5000, 1536}*)
However, you might get better results though by using a more specialized model (and perhaps save a few pesos by using your local compute). SciBert is a BERT-style (bidirectional, masking) language model that has been trained on scientific papers with special tokens for scientific words; this would be a natural choice for computing embeddings of the titles and abstracts for our project. SciBert returns embeddings for each word, so we pool the last layer to get an embedding for the entire text chunk:
scibert = NetAppend[
NetModel["SciBERT Trained on Semantic Scholar Data"],
"pooling" -> SequenceLastLayer[]]
Aside: AllenAI has a special model (SPECTER) for generating document level scientific document embeddings; SentenceTransformers has a python implementation of using this, as well as other tools. These might be a more robust choice for production, but it was harder to pull a pre-trained model while hacking this out.
The general idea is to apply this model to our text to generate the embeddings:
AbsoluteTiming[
scibertEmbeddings = scibert /@ Lookup[articles, "title"];
]
(*{893.686, Null}*)
Note that the typical BERT model has a limit of 512 tokens (about 400 words), and there may be abstracts that are longer than this. The default BERT implementation just discards tokens that exceed this limit (and this is the behavior in the SciBert implementation we are using). But let’s just start with the titles for now. It is pretty slow to process these 5000 records running SciBERT on CPU on my laptop, but this could be sped up by using a machine with a GPU (or even just a faster laptop).
Comment: Many articles are very far removed from chemistry (e.g., German theology papers), so it may be useful to use a very cheap model just to throw away the ones that are obviously not about chemistry. So it goes.
To avoid having to recompute these, let’s save them to disk:
DumpSave["openAIEmbeddings.mx", openAITitleEmbeddings];
DumpSave["scibertEmbeddings.mx", scibertEmbeddings];
(*Reload them in as necessary*)
<< "openAIEmbeddings.mx"
<< "scibertEmbeddings.mx"
We also need a list of titles that are about our separations. I exported a list of articles that I had in Zotero so I could pull our articles that I care about. You may have a more comprehensive list, and you should probably use that.
titles = Normal@Import["~/Downloads/Exported Items.csv", "Dataset", HeaderLines -> 1][[All, "Title"]];
Short[titles]

Just like above, we compute the embeddings for these titles:
referenceTitlesOpenAI = ServiceExecute["OpenAI", "Embedding", {"Input" -> titles}]["data"][[All, "embedding"]];
referenceTitlesScibert = scibert /@ titles;
Dimensions[referenceTitlesScibert]
(*{29, 768}*)
Find the nearest reference articles
The general idea is to compute the CosineDistance between the embedding vectors; closer vectors should be more semantically related. To start, determine how far apart vectors are within our set of reference titles:
Map[
Max@Flatten@DistanceMatrix[#, DistanceFunction -> CosineDistance] &, {referenceTitlesOpenAI, referenceTitlesScibert}]
(*{0.28674, 0.425434}*)
Now define a distance function and find the nearest items (returning the value of the distances):
f = Nearest[referenceTitlesScibert -> "Distance", DistanceFunction -> CosineDistance]
scibertDistances = Map[First]@f@scibertEmbeddings;
Histogram[%]

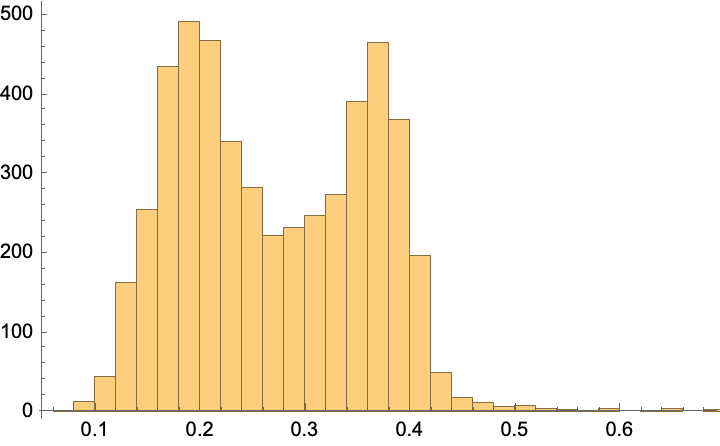
How well did it work? Here are the closest articles:
Dataset@Pick[articles, LessEqualThan[0.1] /@ scibertDistances]
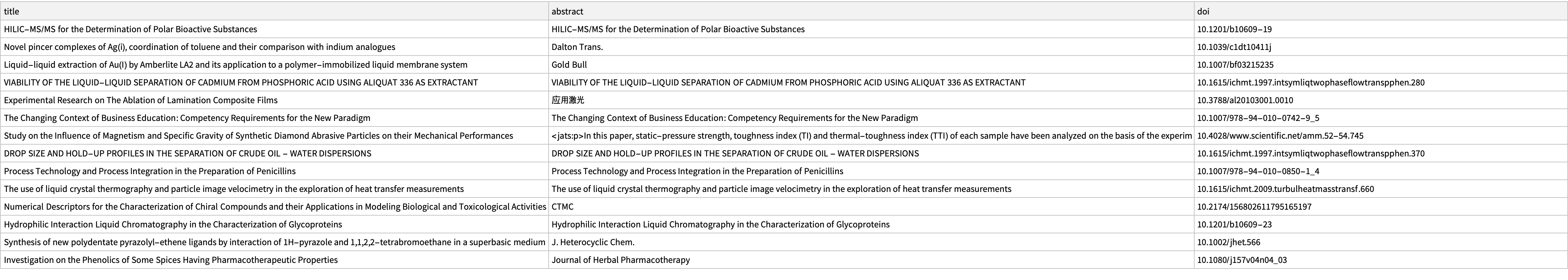
Comment: It certainly seems to be selecting science-y projects and those that have a separation or extraction aspect. But there are many duds too (like an article on business education?)
Would the OpenAI embeddings do better?
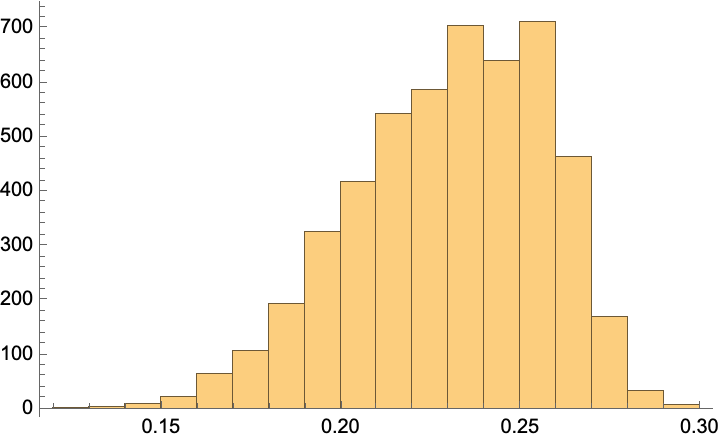
f = Nearest[referenceTitlesOpenAI -> "Distance", DistanceFunction -> CosineDistance];
distances = Map[First]@f@openAITitleEmbeddings;
Histogram[%, PlotRange -> All]
Dataset@Pick[articles, LessEqualThan[0.15] /@ %%]
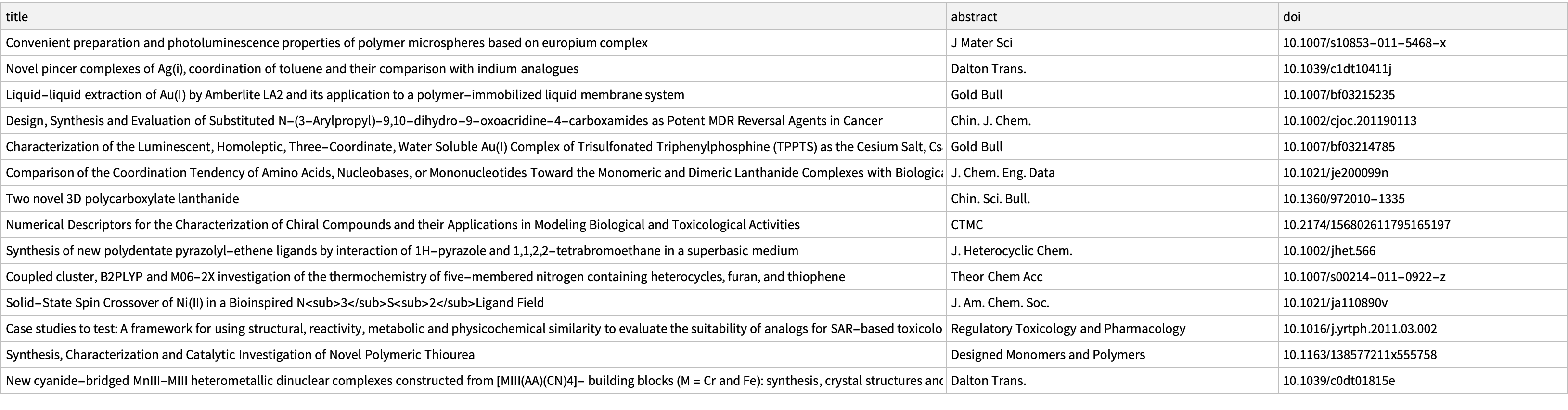
This certainly seems to be more uniformly about chemistry topics, although the relevance of some of these to f-element separation seems limited.
Learnings and next-steps (towards a production system)
-
Data needs:
-
A bigger list of known separation papers (with titles) may be helpful: We can pull these (by DOI) from our various datasets, entered manually, from IDeaL, etc.
- Alternatively, you might try a HYDE-like strategy and just have your favorite GPT-X model generate a list of a few hundred titles for f-element separations.
-
Several examples of data structure irregularity (missing titles, non-English titles, etc.) demonstrated above. Keep this in mind.
-
Titles are generally OK, but abstracts will be better for discriminating between topics. We probably want to use abstract embeddings when available. But MOST of our entries do NOT have an abstract, so this will be limited. So either just give up on abstracts or assume they will be limited.
-
-
The importance of pre-screening: Embeddings are relatively slow and costly, and we have millions of items to process, so it may be useful to discard items that are obviously irrelevant.
-
Even though not all articles have a subject categorization, when it is present, we can use this to pre-screen articles (default to keeping articles in); the goal here is just to reject papers that are obviously not separations
-
It may be possible to also screen the journals (have a white list/black list of journals?)
-
Consider a pre-screening based on titles (must contain at least one keyword)
-
-
Model improvements: The typical approach is a cosine distance, but you may do better with a learned distance function:
-
A simple way to do this might be to train a Siamese network (using a NetPairEmbeddingOperator) to learn a distance function. Consider positive examples to be within our list of known separation papers and negative examples to just be two random papers.
-
Alternatively just use some off-the-shelf Classify model.
-
-
Take a multi-stage approach:
- In addition to a keyword-based pre-screening, use the embedding comparison as a second round of pre-screening. Then start pulling PDFs and downloading abstracts from publisher sites for use with SPECTER or other document-based models.
Elsewhere in the literature…
- Kumar et al. (2023) recently described the user of a BERT-based classifier for identifying relevant recycling literature. Their literature review in the introduction also describes a number of specialized scientific terminology BERT models. They do keyword searches in the Elsevier database, followed by using their BERT-based supervised classifier to identify relevant articles. (They subsequently use this for other data extraction tasks)
- Kedar & co. (2023) text parsing for ternary chalcogenides with GPT3.5
ToJekyll["Semantic Article Retrieval from CrossRef with LLM Embeddings", "science llm machinelearning ml mathematica"]
Other gleanings
- (21 Dec 2023) There is now a SentenceBERTEmbeddings convenience function in the repository